- Privacy Policy
Buy Me a Coffee

Home » Critical Analysis – Types, Examples and Writing Guide

Critical Analysis – Types, Examples and Writing Guide
Table of Contents

Critical Analysis
Definition:
Critical analysis is a process of examining a piece of work or an idea in a systematic, objective, and analytical way. It involves breaking down complex ideas, concepts, or arguments into smaller, more manageable parts to understand them better.
Types of Critical Analysis
Types of Critical Analysis are as follows:
Literary Analysis
This type of analysis focuses on analyzing and interpreting works of literature , such as novels, poetry, plays, etc. The analysis involves examining the literary devices used in the work, such as symbolism, imagery, and metaphor, and how they contribute to the overall meaning of the work.
Film Analysis
This type of analysis involves examining and interpreting films, including their themes, cinematography, editing, and sound. Film analysis can also include evaluating the director’s style and how it contributes to the overall message of the film.
Art Analysis
This type of analysis involves examining and interpreting works of art , such as paintings, sculptures, and installations. The analysis involves examining the elements of the artwork, such as color, composition, and technique, and how they contribute to the overall meaning of the work.
Cultural Analysis
This type of analysis involves examining and interpreting cultural artifacts , such as advertisements, popular music, and social media posts. The analysis involves examining the cultural context of the artifact and how it reflects and shapes cultural values, beliefs, and norms.
Historical Analysis
This type of analysis involves examining and interpreting historical documents , such as diaries, letters, and government records. The analysis involves examining the historical context of the document and how it reflects the social, political, and cultural attitudes of the time.
Philosophical Analysis
This type of analysis involves examining and interpreting philosophical texts and ideas, such as the works of philosophers and their arguments. The analysis involves evaluating the logical consistency of the arguments and assessing the validity and soundness of the conclusions.
Scientific Analysis
This type of analysis involves examining and interpreting scientific research studies and their findings. The analysis involves evaluating the methods used in the study, the data collected, and the conclusions drawn, and assessing their reliability and validity.
Critical Discourse Analysis
This type of analysis involves examining and interpreting language use in social and political contexts. The analysis involves evaluating the power dynamics and social relationships conveyed through language use and how they shape discourse and social reality.
Comparative Analysis
This type of analysis involves examining and interpreting multiple texts or works of art and comparing them to each other. The analysis involves evaluating the similarities and differences between the texts and how they contribute to understanding the themes and meanings conveyed.
Critical Analysis Format
Critical Analysis Format is as follows:
I. Introduction
- Provide a brief overview of the text, object, or event being analyzed
- Explain the purpose of the analysis and its significance
- Provide background information on the context and relevant historical or cultural factors
II. Description
- Provide a detailed description of the text, object, or event being analyzed
- Identify key themes, ideas, and arguments presented
- Describe the author or creator’s style, tone, and use of language or visual elements
III. Analysis
- Analyze the text, object, or event using critical thinking skills
- Identify the main strengths and weaknesses of the argument or presentation
- Evaluate the reliability and validity of the evidence presented
- Assess any assumptions or biases that may be present in the text, object, or event
- Consider the implications of the argument or presentation for different audiences and contexts
IV. Evaluation
- Provide an overall evaluation of the text, object, or event based on the analysis
- Assess the effectiveness of the argument or presentation in achieving its intended purpose
- Identify any limitations or gaps in the argument or presentation
- Consider any alternative viewpoints or interpretations that could be presented
- Summarize the main points of the analysis and evaluation
- Reiterate the significance of the text, object, or event and its relevance to broader issues or debates
- Provide any recommendations for further research or future developments in the field.
VI. Example
- Provide an example or two to support your analysis and evaluation
- Use quotes or specific details from the text, object, or event to support your claims
- Analyze the example(s) using critical thinking skills and explain how they relate to your overall argument
VII. Conclusion
- Reiterate your thesis statement and summarize your main points
- Provide a final evaluation of the text, object, or event based on your analysis
- Offer recommendations for future research or further developments in the field
- End with a thought-provoking statement or question that encourages the reader to think more deeply about the topic
How to Write Critical Analysis
Writing a critical analysis involves evaluating and interpreting a text, such as a book, article, or film, and expressing your opinion about its quality and significance. Here are some steps you can follow to write a critical analysis:
- Read and re-read the text: Before you begin writing, make sure you have a good understanding of the text. Read it several times and take notes on the key points, themes, and arguments.
- Identify the author’s purpose and audience: Consider why the author wrote the text and who the intended audience is. This can help you evaluate whether the author achieved their goals and whether the text is effective in reaching its audience.
- Analyze the structure and style: Look at the organization of the text and the author’s writing style. Consider how these elements contribute to the overall meaning of the text.
- Evaluate the content : Analyze the author’s arguments, evidence, and conclusions. Consider whether they are logical, convincing, and supported by the evidence presented in the text.
- Consider the context: Think about the historical, cultural, and social context in which the text was written. This can help you understand the author’s perspective and the significance of the text.
- Develop your thesis statement : Based on your analysis, develop a clear and concise thesis statement that summarizes your overall evaluation of the text.
- Support your thesis: Use evidence from the text to support your thesis statement. This can include direct quotes, paraphrases, and examples from the text.
- Write the introduction, body, and conclusion : Organize your analysis into an introduction that provides context and presents your thesis, a body that presents your evidence and analysis, and a conclusion that summarizes your main points and restates your thesis.
- Revise and edit: After you have written your analysis, revise and edit it to ensure that your writing is clear, concise, and well-organized. Check for spelling and grammar errors, and make sure that your analysis is logically sound and supported by evidence.
When to Write Critical Analysis
You may want to write a critical analysis in the following situations:
- Academic Assignments: If you are a student, you may be assigned to write a critical analysis as a part of your coursework. This could include analyzing a piece of literature, a historical event, or a scientific paper.
- Journalism and Media: As a journalist or media person, you may need to write a critical analysis of current events, political speeches, or media coverage.
- Personal Interest: If you are interested in a particular topic, you may want to write a critical analysis to gain a deeper understanding of it. For example, you may want to analyze the themes and motifs in a novel or film that you enjoyed.
- Professional Development : Professionals such as writers, scholars, and researchers often write critical analyses to gain insights into their field of study or work.
Critical Analysis Example
An Example of Critical Analysis Could be as follow:
Research Topic:
The Impact of Online Learning on Student Performance
Introduction:
The introduction of the research topic is clear and provides an overview of the issue. However, it could benefit from providing more background information on the prevalence of online learning and its potential impact on student performance.
Literature Review:
The literature review is comprehensive and well-structured. It covers a broad range of studies that have examined the relationship between online learning and student performance. However, it could benefit from including more recent studies and providing a more critical analysis of the existing literature.
Research Methods:
The research methods are clearly described and appropriate for the research question. The study uses a quasi-experimental design to compare the performance of students who took an online course with those who took the same course in a traditional classroom setting. However, the study may benefit from using a randomized controlled trial design to reduce potential confounding factors.
The results are presented in a clear and concise manner. The study finds that students who took the online course performed similarly to those who took the traditional course. However, the study only measures performance on one course and may not be generalizable to other courses or contexts.
Discussion :
The discussion section provides a thorough analysis of the study’s findings. The authors acknowledge the limitations of the study and provide suggestions for future research. However, they could benefit from discussing potential mechanisms underlying the relationship between online learning and student performance.
Conclusion :
The conclusion summarizes the main findings of the study and provides some implications for future research and practice. However, it could benefit from providing more specific recommendations for implementing online learning programs in educational settings.
Purpose of Critical Analysis
There are several purposes of critical analysis, including:
- To identify and evaluate arguments : Critical analysis helps to identify the main arguments in a piece of writing or speech and evaluate their strengths and weaknesses. This enables the reader to form their own opinion and make informed decisions.
- To assess evidence : Critical analysis involves examining the evidence presented in a text or speech and evaluating its quality and relevance to the argument. This helps to determine the credibility of the claims being made.
- To recognize biases and assumptions : Critical analysis helps to identify any biases or assumptions that may be present in the argument, and evaluate how these affect the credibility of the argument.
- To develop critical thinking skills: Critical analysis helps to develop the ability to think critically, evaluate information objectively, and make reasoned judgments based on evidence.
- To improve communication skills: Critical analysis involves carefully reading and listening to information, evaluating it, and expressing one’s own opinion in a clear and concise manner. This helps to improve communication skills and the ability to express ideas effectively.
Importance of Critical Analysis
Here are some specific reasons why critical analysis is important:
- Helps to identify biases: Critical analysis helps individuals to recognize their own biases and assumptions, as well as the biases of others. By being aware of biases, individuals can better evaluate the credibility and reliability of information.
- Enhances problem-solving skills : Critical analysis encourages individuals to question assumptions and consider multiple perspectives, which can lead to creative problem-solving and innovation.
- Promotes better decision-making: By carefully evaluating evidence and arguments, critical analysis can help individuals make more informed and effective decisions.
- Facilitates understanding: Critical analysis helps individuals to understand complex issues and ideas by breaking them down into smaller parts and evaluating them separately.
- Fosters intellectual growth : Engaging in critical analysis challenges individuals to think deeply and critically, which can lead to intellectual growth and development.
Advantages of Critical Analysis
Some advantages of critical analysis include:
- Improved decision-making: Critical analysis helps individuals make informed decisions by evaluating all available information and considering various perspectives.
- Enhanced problem-solving skills : Critical analysis requires individuals to identify and analyze the root cause of a problem, which can help develop effective solutions.
- Increased creativity : Critical analysis encourages individuals to think outside the box and consider alternative solutions to problems, which can lead to more creative and innovative ideas.
- Improved communication : Critical analysis helps individuals communicate their ideas and opinions more effectively by providing logical and coherent arguments.
- Reduced bias: Critical analysis requires individuals to evaluate information objectively, which can help reduce personal biases and subjective opinions.
- Better understanding of complex issues : Critical analysis helps individuals to understand complex issues by breaking them down into smaller parts, examining each part and understanding how they fit together.
- Greater self-awareness: Critical analysis helps individuals to recognize their own biases, assumptions, and limitations, which can lead to personal growth and development.
About the author
Muhammad Hassan
Researcher, Academic Writer, Web developer
You may also like

Cluster Analysis – Types, Methods and Examples

Data Collection – Methods Types and Examples

Delimitations in Research – Types, Examples and...

Discriminant Analysis – Methods, Types and...

Research Process – Steps, Examples and Tips

Research Design – Types, Methods and Examples
An official website of the United States government
The .gov means it’s official. Federal government websites often end in .gov or .mil. Before sharing sensitive information, make sure you’re on a federal government site.
The site is secure. The https:// ensures that you are connecting to the official website and that any information you provide is encrypted and transmitted securely.
- Publications
- Account settings
Preview improvements coming to the PMC website in October 2024. Learn More or Try it out now .
- Advanced Search
- Journal List
- Dtsch Arztebl Int
- v.106(7); 2009 Feb
Critical Appraisal of Scientific Articles
Jean-baptist du prel.
1 Institut für Medizinische Biometrie, Epidemiologie und Informatik (IMBEI), Johannes Gutenberg-Universität, Mainz
Bernd Röhrig
Maria blettner, introduction.
In the era of evidence-based medicine, one of the most important skills a physician needs is the ability to analyze scientific literature critically. This is necessary to keep medical knowledge up to date and to ensure optimal patient care. The aim of this paper is to present an accessible introduction into critical appraisal of scientific articles.
Using a selection of international literature, the reader is introduced to the principles of critical reading of scientific articles in medicine. For the sake of conciseness, detailed description of statistical methods is omitted.
Widely accepted principles for critically appraising scientific articles are outlined. Basic knowledge of study design, structuring of an article, the role of different sections, of statistical presentations as well as sources of error and limitation are presented. The reader does not require extensive methodological knowledge. As far as necessary for critical appraisal of scientific articles, differences in research areas like epidemiology, clinical, and basic research are outlined. Further useful references are presented.
Basic methodological knowledge is required to select and interpret scientific articles correctly.
Despite the increasing number of scientific publications, many physicians find themselves with less and less time to read what others have written. Selection, reading, and critical appraisal of publications is, however, necessary to stay up to date in one’s field. This is also demanded by the precepts of evidence-based medicine ( 1 , 2 ).
Besides the medical content of a publication, its interpretation and evaluation also require understanding of the statistical methodology. Sadly, not even in science are all terms always used correctly. The word "significance," for example, has been overused because significant (or positive) results are easier to get published ( 3 , 4 ).
The aim of this article is to present the essential principles of the evaluation of scientific publications. With the exception of a few specific features, these principles apply equally to experimental, clinical, and epidemiological studies. References to more detailed literature are provided.
Decision making
Before starting a scientific article, the reader must be clear as to his/her intentions. For quick information on a given subject, he/she is advised to read a recent review of some sort, whether a (simple) review article, a systematic review, or a meta-analysis.
The references in review articles point the reader towards more detailed information on the topic concerned. In the absence of any recent reviews on the desired theme, databases such as PubMed have to be consulted.
Regular perusal of specialist journals is an obvious way of keeping up to date. The article title and abstract help the reader to decide whether the article merits closer attention. The title gives the potential reader a concise, accurate first impression of the article’s content. The abstract has the same basic structure as the article and renders the essential points of the publication in greatly shortened form. Reading the abstract is no substitute for critically reading the whole article, but shows whether the authors have succeeded in summarizing aims, methods, results, and conclusions.
The structure of scientific publications
The structure of scientific articles is essentially always the same. The title, summary and key words are followed by the main text. This is divided into Introduction, Methods, Results and Discussion (IMRAD), ending when appropriate with Conclusions and References. The content and purpose of the individual sections are described in detail below.
The Introduction sets out to familiarize the reader with the subject matter of the investigation. The current state of knowledge should be presented with reference to the recent literature and the necessity of the study should be clearly laid out. The findings of the studies cited should be given in detail, quoting numerical results. Inexact phrases such as "inconsistent findings," "somewhat better" and so on are to be avoided. Overall, the text should give the impression that the author has read the articles cited. In case of doubt the reader is recommended to consult these publications him-/herself. A good publication backs up its central statements with references to the literature.
Ideally, this section should progress from the general to the specific. The introduction explains clearly what question the study is intended to answer and why the chosen design is appropriate for this end.
This important section bears a certain resemblance to a cookbook. The description of the procedures should give the reader "recipes" that can be followed to repeat the study. Here are found the essential data that permit appraisal of the study’s validity ( 6 ). The methods section can be divided into subsections with their own headings; for example, laboratory techniques can be described separately from statistical methods.
The methods section should describe all stages of planning, the composition of the study sample (e.g., patients, animals, cell lines), the execution of the study, and the statistical methods: Was a study protocol written before the study commenced? Was the investigation preceded by a pilot study? Are location and study period specified? It should be stated in this section that the study was carried out with the approval of the appropriate ethics committee. The most important element of a scientific investigation is the study design. If for some reason the design is unacceptable, then so is the article, regardless of how the data were analyzed ( 7 ).
The choice of study design should be explained and depicted in clear terms. If important aspects of the methodology are left undescribed, the reader is advised to be wary. If, for example, the method of randomization is not specified, as is often the case ( 8 ), one ought not to assume that randomization took place at all ( 7 ). The statistical methods should be lucidly portrayed and complex statistical parameters and procedures described clearly, with references to the specialist literature. Box 1 contains further questions that may be helpful in evaluation of the Methods section.
Questions on methodology
- Is the study design suited to fulfill the aims of the study?
- Is it stated whether the study is confirmatory, exploratory or descriptive in nature?
- What type of study was chosen, and does it permit the aims of the study to be addressed?
- Is the study’s endpoint precisely defined?
Do epidemiological studies, for instance, give the incidence (rate of new cases), prevalence (current number of cases), mortality (proportion of the population that dies of the disease concerned), lethality (proportion of those with the disease who die of it) or the hospital admission rate (proportion of the population admitted to hospital because of the disease)?
- Are the geographical area, the population, the study period (including duration of follow-up), and the intervals between investigations described in detail?
Study design and implementation are described by Altman ( 7 ), Trampisch and Windeler ( 9 ), and Klug et al. ( 10 ). In experimental studies, precise depiction of the design and execution is vital. The accuracy of a method, i.e. its reliability (precision) and validity (correctness), must be stated. The explanatory power of the results of a clinical study is improved by the inclusion of a control group (active, historical, or placebo controls) and by the randomized assignment of patients to the different arms of the study. The quality can also be raised by blinding of the investigators, which guarantees identical treatment and observation of all study participants. A clinical study should as a rule include an estimation of the required number of patients (case number planning) before the beginning of the study. More detail on clinical studies can be found, for instance, in the book by Schumacher and Schulgen ( 11 ). International recommendations specially formulated for the reporting of randomized, controlled clinical trials are presented in the most recent version of the CONSORT Statement (Consolidated Standards of Reporting Trials) ( 12 ).
Epidemiological investigations can be divided into intervention studies, cohort studies, case-control studies, cross-sectional studies, and ecological studies. Table 1 outlines what type of study is best suited to what situation ( 13 ). One characteristic of a good publication is a precise account of inclusion and exclusion criteria. How high was the response rate (≥80% is good, ≤30% means no or only slight power), and how high was the rate of loss to follow-up, e.g. when participants move away or withdraw their cooperation? To determine whether participants differ from nonparticipants, data on the latter should be included. The selection criteria and the rates of loss to follow-up permit conclusions as to whether the study sample is representative of the target population. A good study description includes information on missing values. Particularly in case-control studies, but also in nonrandomized clinical studies and cohort studies, the choice of the controls must be described precisely. Only then can one be sure that the control group is comparable with the study group and shows no systematic discrepancies that can lead to misinterpretation (confounding) or other problems ( 13 ).
Is it explained how measurements were conducted? Are the instruments and techniques, e.g. measuring devices, scale of measured values, laboratory data, and time point, described in sufficient detail? Were the measurements made under standardized—and thus comparable—conditions in all patients? Details of measurement procedures are important for assessment of accuracy (reliability, validity). The reader must see on what kind of scale the variables are being measured (e.g. eye color, nominal; tumor stage, ordinal; bodyweight, metric), because the type of scale determines what kind of analysis is possible. Descriptive analysis employs descriptive measured values and graphic and/or tabular presentations, whereas in statistical analysis the choice of test has to be taken into consideration. The interpretation and power of the results is also influenced by the scale type. For example, data on an ordinal scale should not be expressed in terms of mean values.
Was there a careful power calculation before the study started? If the number of cases is too low, a real difference, e.g. between the effects of two medications or in the risk of disease in the presence vs. absence of a given environmental factor, may not be detected. One then speaks of insufficient power.
In this section the findings should be presented clearly and objectively, i.e. without interpretation. The interpretation of the results belongs in the ensuing discussion. The results section should address directly the aims of the study and be presented in a well-structured, readily understandable and consistent manner. The findings should first be formulated descriptively, stating statistical parameters such as case numbers, mean values, measures of variation, and confidence intervals. This section should include a comprehensive description of the study population. A second, analytic subsection describes the relationship between characteristics, or estimates the effect of a risk factor, say smoking behavior, on a dependent variable, say lung cancer, and may include calculation of appropriate statistical models.
Besides information on statistical significance in the form of p values, comprehensive description of the data and details on confidence intervals and effect sizes are strongly recommended ( 14 , 15 , 16 ). Tables and figures may improve the clarity, and the data therein should be self-explanatory.
In this section the author should discuss his/her results frankly and openly. Regardless of the study type, there are essentially two goals:
Comparison of the findings with the status quo— The Discussion should answer the following questions: How has the study added to the body of knowledge on the given topic? What conclusions can be drawn from the results? Will the findings of the study lead the author to reconsider or change his/her own professional behavior, e.g. to modify a treatment or take previously unconsidered factors into account? Do the findings suggest further investigations? Does the study raise new, hitherto unanswered questions? What are the implications of the results for science, clinical routine, patient care, and medical practice? Are the findings in accord with those of the majority of earlier studies? If not, why might that be? Do the results appear plausible from the biological or medical viewpoint?
Critical analysis of the study’s limitations— Might sources of bias, whether random or systematic in nature, have affected the results? Even with painstaking planning and execution of the study, errors cannot be wholly excluded. There may, for instance, be an unexpectedly high rate of loss to follow-up (e.g. through patients moving away or refusing to participate further in the study). When comparing groups one should establish whether there is any intergroup difference in the composition of participants lost to follow-up. Such a discrepancy could potentially conceal a true difference between the groups, e.g. in a case-control study with regard to a risk factor. A difference may also result from positive selection of the study population. The Discussion must draw attention to any such differences and describe the patients who do not complete the study. Possible distortion of the study results by missing values should also be discussed.
Systematic errors are particularly common in epidemiological studies, because these are mostly observational rather than experimental in nature. In case-control studies, a typical source of error is the retrospective determination of the study participants’ exposure. Their memories may not be accurate (recall bias). A frequent source of error in cohort studies is confounding. This occurs when two closely connected risk factors are both associated with the dependent variable. Errors of this type can be corrected and revealed by adjustment for the confounding factor. For instance, the fact that smokers drink more coffee than average could lead to the erroneous assumption that drinking coffee causes lung cancer. If potential confounders are not mentioned in the publication, the critical reader should wonder whether the results might not be invalidated by this type of error. If possible confounding factors were not included in the analysis, the potential sources of error should at least be critically debated. Detailed discussion of sources of error and means of correction can be found in the books by Beaglehole and Webb ( 17 , 18 ).
Results that do not attain statistical significance must also be published. Unfortunately, greater importance is still often attached to significant results, so that they are more likely to be published than nonsignificant findings. This publication bias leads to systematic distortions in the body of scientific knowledge. According to a recent review this is particularly true for clinical studies ( 3 ). Only when all valid results of a well-planned and correctly conducted study are published can useful conclusions be drawn regarding the effect of a risk factor on the occurrence of a disease, the value of a diagnostic procedure, the properties of a substance, or the success of an intervention, e.g. a treatment. The investigator and the journal publishing the article are thus obliged to ensure that decisions on important issues can be taken in full knowledge of all valid, scientifically substantiated findings.
It should not be forgotten that statistical significance, i.e. the minimization of the likelihood of a chance result, is not the same as clinical relevance. With a large enough sample, even minuscule differences can become statistically significant, but the findings are not automatically relevant ( 13 , 19 ). This is true both for epidemiological studies, from the public health perspective, and for clinical studies, from the clinical perspective. In both cases, careful economic evaluation is required to decide whether to modify or retain existing practices. At the population level one must ask how often the investigated risk factor really occurs and whether a slight increase in risk justifies wide-ranging public health interventions. From the clinical viewpoint, it must be carefully considered whether, for example, the slightly greater efficacy of a new preparation justifies increased costs and possibly a higher incidence of side effects. The reader has to appreciate the difference between statistical significance and clinical relevance in order to evaluate the results properly.
Conclusions
The authors should concentrate on the most important findings. A crucial question is whether the interpretations follow logically from the results. One should avoid conclusions that are supported neither by one’s own data nor by the findings of others. It is wrong to refer to an exploratory data analysis as a proof. Even in confirmatory studies, one’s own results should, for the sake of consistency, always be considered in light of other investigators’ findings. When assessing the results and formulating the conclusions, the weaknesses of the study must be given due consideration. The study can attain objectivity only if the possibility of erroneous or chance results is admitted. The inclusion of nonsignificant results contributes to the credibility of the study. "Not significant" should not be confused with "no association." Significant results should be considered from the viewpoint of biological and medical plausibility.
So-called levels of evidence scales, as used in some American journals, can help the reader decide to what extent his/her practice should be affected by the content of a given publication ( 20 ). Until all journals offer recommendations of this kind, the individual physician’s ability to read scientific texts critically will continue to play a decisive role in determining whether diagnostic and therapeutic practice are based on up-to-date medical knowledge.
The references are to be presented in the journal’s standard style. The reference list must include all sources cited in the text, tables and figures of the article. It is important to ensure that the references are up to date, in order to make it clear whether the publication incorporates new knowledge. The references cited should help the reader to explore the topic further.
Acknowledgements and conflict of interest statement
This important section must provide information on any sponsors of the study. Any potential conflicts of interest, financial or otherwise, must be revealed in full ( 21 ).
Table 2 and Box 2 summarize the essential questions which, when answered, will reveal the quality of an article. Not all of these questions apply to every publication or every type of study. Further information on the writing of scientific publications is supplied by Gardner et al. ( 19 ), Altman ( 7 ), and Altman et al. ( 22 ). Gardner et al. ( 23 ), Altman ( 7 ), and the CONSORT Statement ( 12 ) provide checklists to assist the evaluation of the statistical content of medical studies.
Critical questions
- Does the study pose scientifically interesting questions?
- Are statements and numerical data supported by literature citations?
- Is the topic of the study medically relevant?
- Is the study innovative?
- Does the study investigate the predefined study goals?
- Is the study design apt to address the aims and/or hypotheses?
- Did practical difficulties (e.g. in recruitment or loss to follow-up) lead to major compromises in study implementation compared with the study protocol?
- Was the number of missing values too large to permit meaningful analysis?
- Was the number of cases too small and thus the statistical power of the study too low?
- Was the course of the study poorly or inadequately monitored (missing values, confounding, time infringements)?
- Do the data support the authors’ conclusions?
- Do the authors and/or the sponsor of the study have irreconcilable financial or ideological conflicts of interest?
Acknowledgments
Translated from the original German by David Roseveare.
Conflict of interest statement
The authors declare no conflicts of interest as defined by the guidelines of the International Committee of Medical Journal Editors.

Writing a Critical Analysis
What is in this guide, definitions, putting it together, tips and examples of critques.
- Background Information
- Cite Sources
Library Links
- Ask a Librarian
- Library Tutorials
- The Research Process
- Library Hours
- Online Databases (A-Z)
- Interlibrary Loan (ILL)
- Reserve a Study Room
- Report a Problem
This guide is meant to help you understand the basics of writing a critical analysis. A critical analysis is an argument about a particular piece of media. There are typically two parts: (1) identify and explain the argument the author is making, and (2), provide your own argument about that argument. Your instructor may have very specific requirements on how you are to write your critical analysis, so make sure you read your assignment carefully.

Critical Analysis
A deep approach to your understanding of a piece of media by relating new knowledge to what you already know.
Part 1: Introduction
- Identify the work being criticized.
- Present thesis - argument about the work.
- Preview your argument - what are the steps you will take to prove your argument.
Part 2: Summarize
- Provide a short summary of the work.
- Present only what is needed to know to understand your argument.
Part 3: Your Argument
- This is the bulk of your paper.
- Provide "sub-arguments" to prove your main argument.
- Use scholarly articles to back up your argument(s).
Part 4: Conclusion
- Reflect on how you have proven your argument.
- Point out the importance of your argument.
- Comment on the potential for further research or analysis.
- Cornell University Library Tips for writing a critical appraisal and analysis of a scholarly article.
- Queen's University Library How to Critique an Article (Psychology)
- University of Illinois, Springfield An example of a summary and an evaluation of a research article. This extended example shows the different ways a student can critique and write about an article
- Next: Background Information >>
- Last Updated: Feb 14, 2024 4:33 PM
- URL: https://libguides.pittcc.edu/critical_analysis

Write a Critical Review of a Scientific Journal Article
1. identify how and why the research was carried out, 2. establish the research context, 3. evaluate the research, 4. establish the significance of the research.
- Writing Your Critique
Ask Us: Chat, email, visit or call

Video: How to Integrate Critical Voice into Your Literature Review

Video: Note-taking and Writing Tips to Avoid Plagiarism

Get assistance
The library offers a range of helpful services. All of our appointments are free of charge and confidential.
- Book an appointment
Read the article(s) carefully and use the questions below to help you identify how and why the research was carried out. Look at the following sections:
Introduction
- What was the objective of the study?
- What methods were used to accomplish this purpose (e.g., systematic recording of observations, analysis and evaluation of published research, assessment of theory, etc.)?
- What techniques were used and how was each technique performed?
- What kind of data can be obtained using each technique?
- How are such data interpreted?
- What kind of information is produced by using the technique?
- What objective evidence was obtained from the authors’ efforts (observations, measurements, etc.)?
- What were the results of the study?
- How was each technique used to obtain each result?
- What statistical tests were used to evaluate the significance of the conclusions based on numeric or graphic data?
- How did each result contribute to answering the question or testing the hypothesis raised in the introduction?
- How were the results interpreted? How were they related to the original problem (authors’ view of evidence rather than objective findings)?
- Were the authors able to answer the question (test the hypothesis) raised?
- Did the research provide new factual information, a new understanding of a phenomenon in the field, or a new research technique?
- How was the significance of the work described?
- Do the authors relate the findings of the study to literature in the field?
- Did the reported observations or interpretations support or refute observations or interpretations made by other researchers?
These questions were adapted from the following sources: Kuyper, B.J. (1991). Bringing up scientists in the art of critiquing research. Bioscience 41(4), 248-250. Wood, J.M. (2003). Research Lab Guide. MICR*3260 Microbial Adaptation and Development Web Site . Retrieved July 31, 2006.
Once you are familiar with the article, you can establish the research context by asking the following questions:
- Who conducted the research? What were/are their interests?
- When and where was the research conducted?
- Why did the authors do this research?
- Was this research pertinent only within the authors’ geographic locale, or did it have broader (even global) relevance?
- Were many other laboratories pursuing related research when the reported work was done? If so, why?
- For experimental research, what funding sources met the costs of the research?
- On what prior observations was the research based? What was and was not known at the time?
- How important was the research question posed by the researchers?
These questions were adapted from the following sources: Kuyper, B.J. (1991). Bringing up scientists in the art of critiquing research. Bioscience 41(4), 248-250. Wood, J.M. (2003). Research Lab Guide. MICR*3260 Microbial Adaptation and Development Web Site . Retrieved July 31, 2006.
Remember that simply disagreeing with the material is not considered to be a critical assessment of the material. For example, stating that the sample size is insufficient is not a critical assessment. Describing why the sample size is insufficient for the claims being made in the study would be a critical assessment.
Use the questions below to help you evaluate the quality of the authors’ research:
- Does the title precisely state the subject of the paper?
- Read the statement of purpose in the abstract. Does it match the one in the introduction?
Acknowledgments
- Could the source of the research funding have influenced the research topic or conclusions?
- Check the sequence of statements in the introduction. Does all the information lead coherently to the purpose of the study?
- Review all methods in relation to the objective(s) of the study. Are the methods valid for studying the problem?
- Check the methods for essential information. Could the study be duplicated from the methods and information given?
- Check the methods for flaws. Is the sample selection adequate? Is the experimental design sound?
- Check the sequence of statements in the methods. Does all the information belong there? Is the sequence of methods clear and pertinent?
- Was there mention of ethics? Which research ethics board approved the study?
- Carefully examine the data presented in the tables and diagrams. Does the title or legend accurately describe the content?
- Are column headings and labels accurate?
- Are the data organized for ready comparison and interpretation? (A table should be self-explanatory, with a title that accurately and concisely describes content and column headings that accurately describe information in the cells.)
- Review the results as presented in the text while referring to the data in the tables and diagrams. Does the text complement, and not simply repeat data? Are there discrepancies between the results in the text and those in the tables?
- Check all calculations and presentation of data.
- Review the results in light of the stated objectives. Does the study reveal what the researchers intended?
- Does the discussion clearly address the objectives and hypotheses?
- Check the interpretation against the results. Does the discussion merely repeat the results?
- Does the interpretation arise logically from the data or is it too far-fetched?
- Have the faults, flaws, or shortcomings of the research been addressed?
- Is the interpretation supported by other research cited in the study?
- Does the study consider key studies in the field?
- What is the significance of the research? Do the authors mention wider implications of the findings?
- Is there a section on recommendations for future research? Are there other research possibilities or directions suggested?
Consider the article as a whole
- Reread the abstract. Does it accurately summarize the article?
- Check the structure of the article (first headings and then paragraphing). Is all the material organized under the appropriate headings? Are sections divided logically into subsections or paragraphs?
- Are stylistic concerns, logic, clarity, and economy of expression addressed?
These questions were adapted from the following sources: Kuyper, B.J. (1991). Bringing up scientists in the art of critiquing research. Bioscience 41(4), 248-250. Wood, J.M. (2003). Research Lab Guide. MICR*3260 Microbial Adaptation and Development Web Site. Retrieved July 31, 2006.
After you have evaluated the research, consider whether the research has been successful. Has it led to new questions being asked, or new ways of using existing knowledge? Are other researchers citing this paper?
You should consider the following questions:
- How did other researchers view the significance of the research reported by your authors?
- Did the research reported in your article result in the formulation of new questions or hypotheses (by the authors or by other researchers)?
- Have other researchers subsequently supported or refuted the observations or interpretations of these authors?
- Did the research make a significant contribution to human knowledge?
- Did the research produce any practical applications?
- What are the social, political, technological, medical implications of this research?
- How do you evaluate the significance of the research?
To answer these questions, look at review articles to find out how reviewers view this piece of research. Look at research articles and databases like Web of Science to see how other people have used this work. What range of journals have cited this article?
These questions were adapted from the following sources:
Kuyper, B.J. (1991). Bringing up scientists in the art of critiquing research. Bioscience 41(4), 248-250. Wood, J.M. (2003). Research Lab Guide. MICR*3260 Microbial Adaptation and Development Web Site . Retrieved July 31, 2006.
- << Previous: Start Here
- Next: Writing Your Critique >>
- Last Updated: Jan 11, 2024 12:42 PM
- URL: https://guides.lib.uoguelph.ca/WriteCriticalReview
Suggest an edit to this guide
This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License.
Paper Information
- Previous Paper
- Paper Submission
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Author Guidelines
p-ISSN: 2162-9463 e-ISSN: 2162-8467
2014; 4(6): 148-155
doi:10.5923/j.edu.20140406.03
A Critical Review Study Conducted on Two Academic Articles Published in the Educational Field: From a Research Prospective
Ivan Hasan Murad
Department of English Language/University of Zakho-Kurdistan Region, Iraq
Copyright © 2014 Scientific & Academic Publishing. All Rights Reserved.
The primary focus of this study is to critically analyse two academic papers published in the educational field in terms of the validity and reliability of their methods of data collection and analysis, research design, and ethical implications. This is done in an attempt to demonstrate the valid procedure of conducting a research paper as a general aim for the current study. This is a desk research study conducted primarily for educational purposes. Data was collected from different resources found in the library of the University of Huddersfield in the United Kingdom. The analysis of the current research was conducted in the light of many educational resources specialized in research papers and publication. Results from the current study show that due to the lack of many standards, Brown's research is not reliable, valid and authentic, whereas Ornprapat and Saovapa's research is outstanding, valid, reliable, and authentic.
Keywords: Critical analysis, Validity, Reliability, Triangulation, Ethical consideration, Sampling
Cite this paper: Ivan Hasan Murad, A Critical Review Study Conducted on Two Academic Articles Published in the Educational Field: From a Research Prospective, Education , Vol. 4 No. 6, 2014, pp. 148-155. doi: 10.5923/j.edu.20140406.03.
Article Outline
1. introduction, 2. overview of the two articles, 2.1. article x, 2.2. article y, 3. literature review, 4. methodology, 4.1. analysis of the research design, 4.2. methods of data collection, 4.3. validity and reliability of the two articles, 4.4. ethical implications, 5. conclusions.
Critically Analyzing Information Sources: Critical Appraisal and Analysis
- Critical Appraisal and Analysis
Initial Appraisal : Reviewing the source
- What are the author's credentials--institutional affiliation (where he or she works), educational background, past writings, or experience? Is the book or article written on a topic in the author's area of expertise? You can use the various Who's Who publications for the U.S. and other countries and for specific subjects and the biographical information located in the publication itself to help determine the author's affiliation and credentials.
- Has your instructor mentioned this author? Have you seen the author's name cited in other sources or bibliographies? Respected authors are cited frequently by other scholars. For this reason, always note those names that appear in many different sources.
- Is the author associated with a reputable institution or organization? What are the basic values or goals of the organization or institution?
B. Date of Publication
- When was the source published? This date is often located on the face of the title page below the name of the publisher. If it is not there, look for the copyright date on the reverse of the title page. On Web pages, the date of the last revision is usually at the bottom of the home page, sometimes every page.
- Is the source current or out-of-date for your topic? Topic areas of continuing and rapid development, such as the sciences, demand more current information. On the other hand, topics in the humanities often require material that was written many years ago. At the other extreme, some news sources on the Web now note the hour and minute that articles are posted on their site.
C. Edition or Revision
Is this a first edition of this publication or not? Further editions indicate a source has been revised and updated to reflect changes in knowledge, include omissions, and harmonize with its intended reader's needs. Also, many printings or editions may indicate that the work has become a standard source in the area and is reliable. If you are using a Web source, do the pages indicate revision dates?
D. Publisher
Note the publisher. If the source is published by a university press, it is likely to be scholarly. Although the fact that the publisher is reputable does not necessarily guarantee quality, it does show that the publisher may have high regard for the source being published.
E. Title of Journal
Is this a scholarly or a popular journal? This distinction is important because it indicates different levels of complexity in conveying ideas. If you need help in determining the type of journal, see Distinguishing Scholarly from Non-Scholarly Periodicals . Or you may wish to check your journal title in the latest edition of Katz's Magazines for Libraries (Olin Reference Z 6941 .K21, shelved at the reference desk) for a brief evaluative description.
Critical Analysis of the Content
Having made an initial appraisal, you should now examine the body of the source. Read the preface to determine the author's intentions for the book. Scan the table of contents and the index to get a broad overview of the material it covers. Note whether bibliographies are included. Read the chapters that specifically address your topic. Reading the article abstract and scanning the table of contents of a journal or magazine issue is also useful. As with books, the presence and quality of a bibliography at the end of the article may reflect the care with which the authors have prepared their work.
A. Intended Audience
What type of audience is the author addressing? Is the publication aimed at a specialized or a general audience? Is this source too elementary, too technical, too advanced, or just right for your needs?
B. Objective Reasoning
- Is the information covered fact, opinion, or propaganda? It is not always easy to separate fact from opinion. Facts can usually be verified; opinions, though they may be based on factual information, evolve from the interpretation of facts. Skilled writers can make you think their interpretations are facts.
- Does the information appear to be valid and well-researched, or is it questionable and unsupported by evidence? Assumptions should be reasonable. Note errors or omissions.
- Are the ideas and arguments advanced more or less in line with other works you have read on the same topic? The more radically an author departs from the views of others in the same field, the more carefully and critically you should scrutinize his or her ideas.
- Is the author's point of view objective and impartial? Is the language free of emotion-arousing words and bias?
C. Coverage
- Does the work update other sources, substantiate other materials you have read, or add new information? Does it extensively or marginally cover your topic? You should explore enough sources to obtain a variety of viewpoints.
- Is the material primary or secondary in nature? Primary sources are the raw material of the research process. Secondary sources are based on primary sources. For example, if you were researching Konrad Adenauer's role in rebuilding West Germany after World War II, Adenauer's own writings would be one of many primary sources available on this topic. Others might include relevant government documents and contemporary German newspaper articles. Scholars use this primary material to help generate historical interpretations--a secondary source. Books, encyclopedia articles, and scholarly journal articles about Adenauer's role are considered secondary sources. In the sciences, journal articles and conference proceedings written by experimenters reporting the results of their research are primary documents. Choose both primary and secondary sources when you have the opportunity.
D. Writing Style
Is the publication organized logically? Are the main points clearly presented? Do you find the text easy to read, or is it stilted or choppy? Is the author's argument repetitive?
E. Evaluative Reviews
- Locate critical reviews of books in a reviewing source , such as the Articles & Full Text , Book Review Index , Book Review Digest, and ProQuest Research Library . Is the review positive? Is the book under review considered a valuable contribution to the field? Does the reviewer mention other books that might be better? If so, locate these sources for more information on your topic.
- Do the various reviewers agree on the value or attributes of the book or has it aroused controversy among the critics?
- For Web sites, consider consulting this evaluation source from UC Berkeley .
Permissions Information
If you wish to use or adapt any or all of the content of this Guide go to Cornell Library's Research Guides Use Conditions to review our use permissions and our Creative Commons license.
- Next: Tips >>
- Last Updated: Apr 18, 2022 1:43 PM
- URL: https://guides.library.cornell.edu/critically_analyzing

A Critical Analysis and Comparison of two Published Academic Journals in Education Field
- Berivan M. A. Abdullah Dept. of English Language, Faculty of Humanities, University of Zakho, Kurdistan Region – Iraq.
The purpose of this study is to critically analyse and evaluate two selected journal articles. The two articles are research papers in the education field; more specifically both journals are about the usage and implementation of Communicative Language Teaching. For ease of reading, each article is dealt with separately. This paper is to pinpoint their weaknesses and strengths. The two chosen papers are also critiqued from various perspectives such as methods of data collection and analysis, research layout and organization, validity and reliability of the data collected, sampling issue and ethical considerations. Although the titles are very similar, the content was different and the papers are written differently using various instruments for collecting data. This study concluded that both articles to some extend have week points such as the use of instruments and the sampling size which were a very limited size especially in article Y. The ethical issues in both articles to some extend are considered, however, in one of the articles, it is less taken into account.
Author Biography
Berivan m. a. abdullah, dept. of english language, faculty of humanities, university of zakho, kurdistan region – iraq..
Dept. of English Language, Faculty of Humanities, University of Zakho, Kurdistan Region – Iraq.

How to Cite
- Endnote/Zotero/Mendeley (RIS)
Authors who publish with this journal agree to the following terms:
- Authors retain copyright and grant the journal right of first publication with the work simultaneously licensed under a Creative Commons Attribution License [ CC BY-NC-SA 4.0 ] that allows others to share the work with an acknowledgment of the work's authorship and initial publication in this journal.
- Authors are able to enter into separate, additional contractual arrangements for the non-exclusive distribution of the journal's published version of the work, with an acknowledgment of its initial publication in this journal.
- Authors are permitted and encouraged to post their work online.
Similar Articles
- Zaki H. Mohammad, Mohamed B. Al-Azzawi, Kurdish EFL Student Teachers’ Perceptions Towards Their School-Based Teaching Practice Course , Humanities Journal of University of Zakho: Vol. 12 No. 1 (2024): 1, Jan - 30, Mar
- Dilbreen F. Abdullah, Fakher O. Mohammed , Investigating the use of fillers by kurdish efl university students in relation to speaking fluency , Humanities Journal of University of Zakho: Vol. 11 No. 3 (2023): July-September
- Basheer Y. Ismail, Najat W. Khalid, THE EFFECT OF APPLYING IFRS 15 ON THE INCOME STATEMENT: AN APPLY STUDY ON AL-KHAYATE AL- HADYTHA FIRM , Humanities Journal of University of Zakho: Vol. 12 No. 1 (2024): 1, Jan - 30, Mar
You may also start an advanced similarity search for this article.
Information
- For Readers
- For Authors
- For Librarians
How to rigister and create an account
How to make submission
- Administration, legal and financial problems in the iraqi tax system 847
- Strategies of Marketing Mix and Its Role in Increasing of Market Share: Exploring Study for The Opinions of Employees in City Centre Company at Zakho City 751
- The Concept of justice And Its Disciplines With Narrators 721
- Abdelhamid Ben Badis his life and his political and cultural role 1840-1989 648
- The impact of monetary policy variables on Iraq’s recessionary inflation for the period 2004-2021 575
Indexing / Member in

- The Open University
- Guest user / Sign out
- Study with The Open University

My OpenLearn Profile
Personalise your OpenLearn profile, save your favourite content and get recognition for your learning
About this free course
Become an ou student, download this course, share this free course.

Start this free course now. Just create an account and sign in. Enrol and complete the course for a free statement of participation or digital badge if available.
1 Important points to consider when critically evaluating published research papers
Simple review articles (also referred to as ‘narrative’ or ‘selective’ reviews), systematic reviews and meta-analyses provide rapid overviews and ‘snapshots’ of progress made within a field, summarising a given topic or research area. They can serve as useful guides, or as current and comprehensive ‘sources’ of information, and can act as a point of reference to relevant primary research studies within a given scientific area. Narrative or systematic reviews are often used as a first step towards a more detailed investigation of a topic or a specific enquiry (a hypothesis or research question), or to establish critical awareness of a rapidly-moving field (you will be required to demonstrate this as part of an assignment, an essay or a dissertation at postgraduate level).
The majority of primary ‘empirical’ research papers essentially follow the same structure (abbreviated here as IMRAD). There is a section on Introduction, followed by the Methods, then the Results, which includes figures and tables showing data described in the paper, and a Discussion. The paper typically ends with a Conclusion, and References and Acknowledgements sections.
The Title of the paper provides a concise first impression. The Abstract follows the basic structure of the extended article. It provides an ‘accessible’ and concise summary of the aims, methods, results and conclusions. The Introduction provides useful background information and context, and typically outlines the aims and objectives of the study. The Abstract can serve as a useful summary of the paper, presenting the purpose, scope and major findings. However, simply reading the abstract alone is not a substitute for critically reading the whole article. To really get a good understanding and to be able to critically evaluate a research study, it is necessary to read on.
While most research papers follow the above format, variations do exist. For example, the results and discussion sections may be combined. In some journals the materials and methods may follow the discussion, and in two of the most widely read journals, Science and Nature, the format does vary from the above due to restrictions on the length of articles. In addition, there may be supporting documents that accompany a paper, including supplementary materials such as supporting data, tables, figures, videos and so on. There may also be commentaries or editorials associated with a topical research paper, which provide an overview or critique of the study being presented.
Box 1 Key questions to ask when appraising a research paper
- Is the study’s research question relevant?
- Does the study add anything new to current knowledge and understanding?
- Does the study test a stated hypothesis?
- Is the design of the study appropriate to the research question?
- Do the study methods address key potential sources of bias?
- Were suitable ‘controls’ included in the study?
- Were the statistical analyses appropriate and applied correctly?
- Is there a clear statement of findings?
- Does the data support the authors’ conclusions?
- Are there any conflicts of interest or ethical concerns?
There are various strategies used in reading a scientific research paper, and one of these is to start with the title and the abstract, then look at the figures and tables, and move on to the introduction, before turning to the results and discussion, and finally, interrogating the methods.
Another strategy (outlined below) is to begin with the abstract and then the discussion, take a look at the methods, and then the results section (including any relevant tables and figures), before moving on to look more closely at the discussion and, finally, the conclusion. You should choose a strategy that works best for you. However, asking the ‘right’ questions is a central feature of critical appraisal, as with any enquiry, so where should you begin? Here are some critical questions to consider when evaluating a research paper.
Look at the Abstract and then the Discussion : Are these accessible and of general relevance or are they detailed, with far-reaching conclusions? Is it clear why the study was undertaken? Why are the conclusions important? Does the study add anything new to current knowledge and understanding? The reasons why a particular study design or statistical method were chosen should also be clear from reading a research paper. What is the research question being asked? Does the study test a stated hypothesis? Is the design of the study appropriate to the research question? Have the authors considered the limitations of their study and have they discussed these in context?
Take a look at the Methods : Were there any practical difficulties that could have compromised the study or its implementation? Were these considered in the protocol? Were there any missing values and, if so, was the number of missing values too large to permit meaningful analysis? Was the number of samples (cases or participants) too small to establish meaningful significance? Do the study methods address key potential sources of bias? Were suitable ‘controls’ included in the study? If controls are missing or not appropriate to the study design, we cannot be confident that the results really show what is happening in an experiment. Were the statistical analyses appropriate and applied correctly? Do the authors point out the limitations of methods or tests used? Were the methods referenced and described in sufficient detail for others to repeat or extend the study?
Take a look at the Results section and relevant tables and figures : Is there a clear statement of findings? Were the results expected? Do they make sense? What data supports them? Do the tables and figures clearly describe the data (highlighting trends etc.)? Try to distinguish between what the data show and what the authors say they show (i.e. their interpretation).
Moving on to look in greater depth at the Discussion and Conclusion : Are the results discussed in relation to similar (previous) studies? Do the authors indulge in excessive speculation? Are limitations of the study adequately addressed? Were the objectives of the study met and the hypothesis supported or refuted (and is a clear explanation provided)? Does the data support the authors’ conclusions? Maybe there is only one experiment to support a point. More often, several different experiments or approaches combine to support a particular conclusion. A rule of thumb here is that if multiple approaches and multiple lines of evidence from different directions are presented, and all point to the same conclusion, then the conclusions are more credible. But do question all assumptions. Identify any implicit or hidden assumptions that the authors may have used when interpreting their data. Be wary of data that is mixed up with interpretation and speculation! Remember, just because it is published, does not mean that it is right.
O ther points you should consider when evaluating a research paper : Are there any financial, ethical or other conflicts of interest associated with the study, its authors and sponsors? Are there ethical concerns with the study itself? Looking at the references, consider if the authors have preferentially cited their own previous publications (i.e. needlessly), and whether the list of references are recent (ensuring that the analysis is up-to-date). Finally, from a practical perspective, you should move beyond the text of a research paper, talk to your peers about it, consult available commentaries, online links to references and other external sources to help clarify any aspects you don’t understand.
The above can be taken as a general guide to help you begin to critically evaluate a scientific research paper, but only in the broadest sense. Do bear in mind that the way that research evidence is critiqued will also differ slightly according to the type of study being appraised, whether observational or experimental, and each study will have additional aspects that would need to be evaluated separately. For criteria recommended for the evaluation of qualitative research papers, see the article by Mildred Blaxter (1996), available online. Details are in the References.
Activity 1 Critical appraisal of a scientific research paper
A critical appraisal checklist, which you can download via the link below, can act as a useful tool to help you to interrogate research papers. The checklist is divided into four sections, broadly covering:
- some general aspects
- research design and methodology
- the results
- discussion, conclusion and references.
Science perspective – critical appraisal checklist [ Tip: hold Ctrl and click a link to open it in a new tab. ( Hide tip ) ]
- Identify and obtain a research article based on a topic of your own choosing, using a search engine such as Google Scholar or PubMed (for example).
- The selection criteria for your target paper are as follows: the article must be an open access primary research paper (not a review) containing empirical data, published in the last 2–3 years, and preferably no more than 5–6 pages in length.
- Critically evaluate the research paper using the checklist provided, making notes on the key points and your overall impression.
Critical appraisal checklists are useful tools to help assess the quality of a study. Assessment of various factors, including the importance of the research question, the design and methodology of a study, the validity of the results and their usefulness (application or relevance), the legitimacy of the conclusions, and any potential conflicts of interest, are an important part of the critical appraisal process. Limitations and further improvements can then be considered.
Advertisement
A global analysis of habitat fragmentation research in reptiles and amphibians: what have we done so far?
- Review Paper
- Open access
- Published: 08 January 2023
- Volume 32 , pages 439–468, ( 2023 )
Cite this article
You have full access to this open access article
- W. C. Tan ORCID: orcid.org/0000-0002-6067-3528 1 ,
- A. Herrel ORCID: orcid.org/0000-0003-0991-4434 2 , 3 , 4 &
- D. Rödder ORCID: orcid.org/0000-0002-6108-1639 1
7445 Accesses
10 Citations
27 Altmetric
Explore all metrics
Habitat change and fragmentation are the primary causes of biodiversity loss worldwide. Recent decades have seen a surge of funding, published papers and citations in the field as these threats to biodiversity continue to rise. However, how research directions and agenda are evolving in this field remains poorly understood. In this study, we examined the current state of research on habitat fragmentation (due to agriculture, logging, fragmentation, urbanisation and roads) pertaining to two of the most threatened vertebrate groups, reptiles and amphibians. We did so by conducting a global scale review of geographical and taxonomical trends on the habitat fragmentation types, associated sampling methods and response variables. Our analyses revealed a number of biases with existing research efforts being focused on three continents (e.g., North America, Europe and Australia) and a surplus of studies measuring species richness and abundance. However, we saw a shift in research agenda towards studies utilising technological advancements including genetic and spatial data analyses. Our findings suggest important associations between sampling methods and prevalent response variables but not with the types of habitat fragmentation. These research agendas are found homogeneously distributed across all continents. Increased research investment with appropriate sampling techniques is crucial in biodiversity hotpots such as the tropics where unprecedented threats to herpetofauna exist.
Similar content being viewed by others
Habitat conservation research for amphibians: methodological improvements and thematic shifts.
Gentile Francesco Ficetola
Effect of landscape composition and configuration on biodiversity at multiple scales: a case study with amphibians from Sierra Madre del Sur, Oaxaca, Mexico
Daniel G. Ramírez-Arce, Leticia M. Ochoa-Ochoa & Andrés Lira-Noriega

A dry future for the Everglades favors invasive herpetofauna
Hunter J. Howell, Giacomo L. Delgado, … Christopher A. Searcy
Avoid common mistakes on your manuscript.
Introduction
Habitat loss and fragmentation are the predominant causes underlying widespread biodiversity changes in terrestrial ecosystems (Fahrig 2003 ; Newbold et al. 2015 ). These processes may cause population declines by disrupting processes such as dispersal, gene flow, and survival. Over the past 30 years habitat loss and fragmentation have been suggested to have reduced biodiversity by up to 75% in different biomes around the world (Haddad et al. 2015 ). This is mainly due to the clearing of tropical forests, the expansion of agricultural landscapes, the intensification of farmland production, and the expansion of urban areas (FAO and UNEP 2020 ). The rate of deforestation and corresponding land conversions of natural habitats are happening rapidly and will continue to increase in the future at an accelerated rate, particularly in biodiversity hotspots (Deikumah et al. 2014 ; Habel et al. 2019 ; FAO and UNEP 2020 ).
For this reason, habitat fragmentation has been a central research focus for ecologists and conservationists over the past two decades (Fardila et al. 2017 ). However, habitat fragmentation consists of two different processes: loss of habitat and fragmentation of existing habitat (Fahrig 2003 ). The former simply means the removal of habitat, and latter is the transformation of continuous areas into discontinuous patches of a given habitat. In a radical review, Fahrig ( 2003 ) suggested that fragmentation per se, i.e., the breaking up of habitat after controlling for habitat loss, has a weaker or even no effect on biodiversity compared to habitat loss. She further recommended that the effects of these two components should be measured independently (Fahrig 2017 ). Despite being recognised as two different processes, researchers tend not to distinguish between their effects and commonly lump the combined consequences under a single umbrella term “habitat fragmentation” (Fahrig 2003 , 2017 ; Lindenmayer and Fischer 2007 ; Riva and Fahrig 2022 ). Nonetheless, fragmentation has been widely recognised in the literature and describes changes that occur in landscapes, including the loss of habitat (Hadley and Betts 2016 ). Hence, to avoid imprecise or inconsistent use of terminology and provide a holistic view of the effect of modified landscapes, we suggest the term “habitat fragmentation” to indicate any type of landscape change, both habitat loss and fragmentation throughout the current paper.
One main conundrum is that biodiversity decline does not occur homogeneously everywhere nor among all species (Blowes et al. 2019 ). Moreover, we should expect a global disparity in biodiversity responses to habitat fragmentation across different biomes (Newbold et al. 2020 ; Cordier et al. 2021 ). For example, tropical regions are predicted to have higher negative effects of habitat fragmentation than temperate regions. There are two possible reasons: a) higher intensification of land use change in the tropics (Barlow et al. 2018 ), and b) forest animals in the tropics are less likely to cross open areas (Lindell et al. 2007 ). Furthermore, individual species respond to landscape modification differently; some thrive whereas others decline (Fahrig 2003 ). Habitat specialists with broader habitat tolerance and wide-ranging distributions are most likely to benefit from increase landscape heterogeneity and more open and edge habitat (Hamer and McDonnell 2008 ; Newbold et al. 2014 ; Palmeirim et al. 2017 ). Therefore, appropriate response metrics should be used in measuring the effect of habitat fragmentation on biodiversity depending on the taxa group, biome and scale of study as patterns of richness can sometimes be masked by the abundance of generalist species (Riemann et al. 2015 ; Palmeirim et al. 2017 ).
Previous reviews have identified general patterns and responses of reptile and amphibian populations to habitat modification. They have been largely centred around specific types of habitat fragmentation: land use change (Newbold et al. 2020 ), logging (Sodhi et al. 2004 ), fragmentation per se (Fahrig 2017 ), urbanisation (Hamer and McDonnell 2008 ; McDonald et al. 2013 ), fire (Driscoll et al. 2021 ), and roads (Rytwinski and Fahrig 2012 ). Few reviews have, however, attempted a global synthesis of all types of land use changes and even fewer have addressed biases in geographical regions and taxonomical groups (but see Gardner et al. ( 2007 ) and Cordier et al. ( 2021 )). Gardner et al. ( 2007 ) synthesised the extant literature and focused on 112 papers on the consequences of habitat fragmentation on reptiles and amphibians published between 1945 and 2006. They found substantial biases across geographic regions, biomes, types of data collected as well as sampling design and effort. However, failure to report basic statistics by many studies prevented them from performing meta-analyses on research conclusions. More recently, a review by Cordier et al. ( 2021 ) conducted a meta-analysis based on 94 primary studies on the overall effects of land use changes through time and across the globe. Yet, there has been no comprehensive synthesis on the research patterns and agenda of published literature on habitat fragmentation associated with the recent advances of novel research tools and techniques. Therefore, our review may provide new insights of the evolution and biases in the field over the last decades and provide a basis for future research directions. Knowledge gaps caused by these biases could hamper the development of habitat fragmentation research and the implementation of effective strategies for conservation.
We aim to remedy this by examining research patterns for the two vertebrate classes Amphibia and Reptilia, at a global scale. We chose amphibians and reptiles for several reasons. First, habitat fragmentation research has been dominated by birds and mammals (Fardila et al. 2017 ). Reptiles and amphibians, on the other hand, are under-represented; together, they constitute only 10% of the studies (Fardila et al. 2017 ). Second, high proportions of amphibian and reptile species are threatened globally. To date, more than one third of amphibian (40%) and one in five reptile species (21%) are threatened with extinction (Stuart et al. 2004 ; Cox et al. 2022 ). Amphibians are known to be susceptible to land transformation as a result of their cryptic nature, habitat requirements, and reduced dispersal ability (Green 2003 ; Sodhi et al. 2008 ; Ofori‐Boateng et al. 2013 ; Nowakowski et al. 2017 ). Although poorly studied (with one in five species classified as data deficient) (Böhm et al. 2013 ), reptiles face the same threats as those impacting amphibians (Gibbons et al. 2000 ; Todd et al. 2010 ; Cox et al. 2022 ). Reptiles have small distributional ranges with high endemism compared to other vertebrates and as such are likely vulnerable to habitat fragmentation (Todd et al. 2010 ; Meiri et al. 2018 ). Third, both these groups are poikilotherms whose physiology makes them highly dependent on temperature and precipitation levels. Hence, they are very sensitive to changing thermal landscapes (Nowakowski et al. 2017 ).
Here, we first ask how is the published literature distributed across geographic regions and taxa? Is there a bias in the geographic distribution of species studied compared to known species? It is well known that conservation and research efforts are often concentrated in wealthy and English-speaking countries (Fazey et al. 2005 ), but has this bias improved over the years? Second, how are researchers conducting these studies? We assess whether certain sampling methods and response variables are associated to specific types of habitat fragmentation. Over the past decades new tools and techniques are constantly being discovered or developed. Combinations of methodologies are now shedding new light on biodiversity responses and consequences of habitat fragmentation. In particular, genetic techniques are useful in detecting changes in population structure, identifying isolated genetic clusters, and in estimating dispersal (Smith et al. 2016 ). Similarly, habitat occupancy and modelling can also provide powerful insights into dispersal (Driscoll et al. 2014 ). Remote sensing data are now used in analysing effects of area, edge, and isolation (Ray et al. 2002 ). Finally, how are these associations or research agendas distributed across space? We expect to find geographic structure of emerging agendas across the globe. For instance, we predict genetic studies to be located in North America and Europe but also in East Asian countries such as China and Japan as a result of their advancement in genetics (Forero et al. 2016 ). On the other hand, simple biodiversity response indicators which do not require extensive capacity building and application of advanced technologies are likely more used in developing regions of the world (Barber et al. 2014 ). These findings are valuable to evaluate and update the global status of our research on the effects of habitat fragmentation on herpetofauna and to suggest recommendations for conservation plans.
Materials and methods
Data collection.
We conducted the review according to the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines (Fig. 1 ) (Moher et al. 2009 ). We conducted a comprehensive and exhaustive search using Web of Science to review published studies reporting the consequences of habitat fragmentation on amphibians and reptiles. We consulted the database in November 2019 by using two general search strings: (1) Habitat fragmentation AND (frog* OR amphib* OR salamander* OR tadpole*) (2) Habitat fragmentation AND (reptil* OR snake* OR lizard* OR turtle* OR crocodile*). This returned a total of 869 records from search (1) and 795 from search (2), with 1421 unique records remaining after duplicates were removed. We did not include “habitat loss” in our search term as it would only introduce unrelated articles focusing on biodiversity and conservation management instead of methodology and mechanistic approaches.
PRISMA flow-diagram of the study selection process
Throughout, we use the term papers to refer to individual journal article records. Out of the 1421 papers, we were unfortunately not able to locate seven papers from Acta Oecologica, Zoology: Analysis of Complex Systems, Israel Journal of Ecology and Evolution, Western North American Naturalist, Natural Areas Journal, Ecology, and the Herpetological Journal. We screened all articles from the title through the full text to determine whether they met our criteria for inclusion. To be included, studies needed to fulfil several criteria. First, papers needed to be peer-reviewed journal articles containing data collected on reptiles and/or amphibians at the species level (224 articles rejected because no species-specific data was available). Reviews and metastudies (n = 102) were excluded from the data analysis as they may represent duplicates as they are mainly based on data sets from other papers, but these form an integral part of our discussion. Furthermore, papers which do not provide data on contemporary time scales such as long-term (> 10, 000 years ago) changes on the paleo-spatial patterns (n = 59) were excluded. Because the effects of fragmentation per se have been measured inconsistently by many authors and have not been differentiated from habitat removal (Fahrig 2003 ), we consider any recent anthropogenic habitat degradation, and modification at patch or/and landscape scales during the Holocene as an effect of habitat fragmentation. Only papers which examined direct or indirect effects of habitat fragmentation were included in our analysis, regardless of the magnitude and direction. Papers which did not mention specific types of habitat fragmentation as the focus of their study (n = 338) were excluded.
Geographical and taxonomical distribution
Using the selected papers, we compiled a taxonomic and geographical database for each paper: (a) GPS or georeferenced location of the study site; (b) the focal group investigated (amphibian and/or reptile); (c) taxonomic groups (order, family, genus).
We listed the overall number of species studied covered by selected papers in each continent and compared them to the total number of currently described species. We obtained total described species of both reptiles and amphibians from the following sources: ReptileDatabase (Uetz et al. 2021 ) and AmphibiaWeb (AmphibiaWeb 2021 ). Then, we calculated the proportions of species covered by the selected papers compared to total number of described species for each continent. We did not update species nomenclature from selected papers as the mismatches from these potentially outdated taxonomies would be insignificant in our analyses.
Categorisation of papers
Each paper is classified into three main types of data collected: forms of habitat fragmentation, sampling methods, and response variables (Online Appendix 1). A paper can be classified into one or multiple categories in each type of data. The types of data and their following categories were:
Forms of habitat fragmentation
We recorded different types of habitat fragmentation from the selection of studies: (1) “Fragmentation” (includes patch isolation, edge and area effects); (2) “Agriculture” (includes any form of commercial and subsistence farming such as monocultures, plantations, and livestock farming); (3) “Logging” (e.g., agroforestry and silviculture); (4) “Mining” (presence of mining activities); (5) “Urbanisation” (includes presence of cities, towns or villages and parks created for recreational purposes); (6) “Road” (includes any vehicle roadway such as railways and highways) and (7) “Other types of habitat fragmentation” (e.g., fire, river dams, ditches, diseases, desertification etc.). Many studies deal with more than one type of habitat fragmentation. However, we made sure the selection for fragmentation forms is mainly based on the focus and wordings in the methodology section.
Sampling methods
We report trends in the design and sampling methods among the compiled studies over the last three decades. Due to the substantial variability in the level of sample design information reported by different studies, we narrowed them down into six general categories representing common sampling methods. Common methods used in estimating herpetofauna diversity (e.g., visual transect surveys, acoustic monitoring and trapping methods) were not included in the analyses due to their omnipresence in the data. The categories are:
(1) “Genetics” studies documented any use of codominant markers (i.e., allozymes and microsatellites), dominant markers [i.e., DNA sequences, random amplified polymorphic DNA (RAPDs) and amplified fragment length polymorphism (AFLPs)] to analyse genetic variability and gene diversity respectively. (2) “Direct tracking methods” studies measured potential dispersal distances or species movement patterns by means of radio telemetry, mark-recapture methods, or fluorescent powder tracking. (3) “Aerial photographs” studies reported the use of aerial photographs while (4) “GIS/Satellite image” studies described the use of satellite imagery and land cover data (i.e., Landsat) and GIS programs (e.g., QGIS and ArcGIS, etc.) in analysing spatial variables. (5) “Experimental” studies involved predictions tested through empirical studies, regardless if they occur naturally or artificially; in a natural or a captive environment. (6) “Prediction/simulation models” studies made use of techniques such as ecological niche models, habitat suitability (i.e., occurrence and occupancy models) and simulations for probability of survival and population connectivity.
Response variables
To further conceptualise how the effects of habitat fragmentation are measured, we assigned 12 biodiversity or ecological response variables. We recorded the type of data that was used in all selected studies: (1) “Species richness or diversity” which are measures of species richness, evenness or diversity (such as the Shannon–Wiener index) (Colwell 2009 ); (2) “Functional richness or species guilds” describes diversity indices based on functional traits (such as body size, reproductive modes, microhabitat association or taxonomic groups); (3) “Presence/absence” or species occupancy; (4) “Population” includes an estimation of population size or density (only when measured specifically in the paper). It includes genetic variation and divergence within and between populations; (5) “Abundance” or counts of individuals for comparison between habitat fragmentation type or species; (6) “Dispersal” takes into account any displacement or movement and can include indirect measurements of dispersal using genetic techniques; (7) “Breeding sites” which measures available breeding or reproduction sites; (8) “Fitness measure” are records of any physiological, ecological or behavioural changes; (9) “Interspecific interaction” depicts any interaction between species including competition and predation; (10) “Extinction or colonisation rate” counts the number of population extinctions or colonisations within a time period; (11) “Microhabitat preference” includes any direct observation made on an individual’s surrounding environmental features (substrate type, perch height, vegetation type, distance to cover etc.); (12) “Generalist or specialist comparison” involves any comparison made between generalist and specialist species. Generalists are able to thrive in various environments whereas specialists occupy a much narrower niche; (13) “Other response variables” can include road kill mortality counts, infection rate of diseases, injury, or any effect from introduced animals and a variety of other responses.
Data analysis
All statistical analyses were conducted in the open source statistical software package R 4.1.0 (R Core Team 2021 ). To gain a broad insight into our understanding of the complexity of habitat fragmentation we applied a Multiple Correspondence Analysis (MCA) (Roux and Rouanet 2004 ) and Hierarchical Clustering on Principle Components (HCPC) (Ward 1963 ) to investigate potential interactions between forms of habitat fragmentation, sampling methods and response variables. MCA is ideal for investigation of datasets with multiple categorical variables and exploration of unbiased relationships between these variables.
We first separate the dataset into papers concerning amphibians or reptiles. The MCA was performed using the MCA function from FactoMineR package of R version 3.1 (Lê et al. 2008 ). To identify subgroups (cluster) of similar papers within our dataset, we performed cluster analysis on our MCA results using HCPC. The cluster results are then visualised in factor map and dendrogram for easier interpretation using factoextra package. This allows us to identify categorical variables which have the highest effect within each cluster. Statistical analyses were considered significant at α = 0.05, while a p between 0.10 and 0.05 was considered as a tendency. The p-value is less than 5% when one category is significantly linked to other categories. The V tests show whether the category is over-expressed (positive values) or under-expressed (negative values) in the cluster (Lebart et al. 1995 ).
Results from the literature review were also analysed with VOSviewer, freeware for constructing and visualising bibliometric networks ( http://www.vosviewer.com/ ). The program uses clustering techniques to analyse co-authors, co-occurrence of keywords, citations, or co-citations (van Eck and Waltman 2014 ). First, we analyse co-authorships of countries to provide a geographical representation of groups of authors in various countries over the past 30 years. Each circle represents an author’s country and the size represents the collaboration frequency with other countries. The lines between the nodes represent the collaboration networks between the countries while the thickness of the lines indicates the collaboration intensities between them. Lastly, to complement the MCA and HCPC, we used VOSviewer to analyse a clustering solution of categories at an aggregate level. Aggregate clustering is a meta-clustering method to improve the robustness of clustering and does not require a priori information about the number of clusters. In this case, instead of author’s keywords, we used the co-occurrence of categories associated to each selected paper as input to run the software.
We identified a total of 698 papers published between January 1991 and November 2019 reporting consequences of habitat fragmentations corresponding to our selection criteria (Fig. 1 ). The complete list of studies included (hereafter termed “selected papers”) is available in Online Appendix 2. The distribution of these selected papers between focal groups and among continents was non-homogeneous (Fig. 2 ). Selected papers reviewed were predominantly studies which were conducted in North America 310 (44%) and Europe 131 (19%), but also from Oceania 104 (15%), South America 85 (12%), Asia 37 (5%) and Africa 31 (5%). For co-authorships between countries based on VOSviewer, the minimum document number of a country was set as 5 and a total of 21 and 14 countries met the threshold for amphibians and reptiles respectively (Fig. 3 ). For amphibians, countries in the American continent such as United States of America or USA (178 articles), Brazil (38 articles) and Canada (35 articles) have the largest research weight (Fig. 3 a). Authors from the USA have the largest international cooperation network, followed by Brazil. Australia and other European countries such as Germany, France and England also have high collaboration relationships with other countries. In contrast, reptile studies were mainly concentrated around two countries: the USA (139 articles) and Australia (86 articles) (Fig. 3 b). No other country from the rest of the world has more than 20 articles. While both the USA and Australia have the largest collaboration networks, Canada, Spain and Mexico are also highly cooperative with authors from other countries.
Map of study locations for a amphibians and b reptiles with each circle representing the study location of papers included in the review. The colour scale of the continents ranging from 0 – 0.9 indicates the proportions of amphibian and reptile species represented in the reviewed papers when compared to known species in the world (obtained from AmphibiaWeb and ReptileDatabase): a Europe (0.73), Africa (0.23), North America (0.23), South America (0.18), Oceania (0.07) and Asia (0.06) and b Europe (0.27), Oceania (0.18), Africa (0.12), North America (0.11), South America (0.09) and Asia (0.02)
Co-authorship map of countries involved in habitat fragmentation research in a amphibians and b reptiles. The colours represent the continents countries belong to. Each circle represents an author’s country and the size represents the collaboration frequency with other countries. The lines between the nodes represent the collaboration networks between the countries while the thickness of the lines indicates the collaboration intensities between them. Category co-occurrence network maps for c amphibians and d reptiles. The colour represents the different cluster groups each category belongs to. Abbreviations for the categories in forms of habitat change: fragmentation (FGM), agriculture (AGR), Logging (LOG), Mining (MIN), Urbanisation (URB), road (RD), other habitat fragmentation (OHC); sampling methods: genetics (GEN), direct tracking method (DTM), aerial photographs (APT), GIS/ Satellite images (GIS), experimental (EXP), prediction/ simulation models (PSM) and response variables: species richness/ diversity (SPR), functional richness/ species guild (FCR), presence/ absence (PAS), population (POP), abundance (ABD), dispersal (DSP), breeding sites (BRD), fitness measure (FIT), interspecific interaction (INT),extinction/ colonisation rate (ECR), microhabitat preference (MHP), comparison between generalist and specialist (CGS), other response varialbes (ORV) (see also Online Appendix 1). Maps are created in VOSviewer
Overall, over half of all selected papers included only amphibians (376 papers; 54%), whilst 276 papers (39%) included only reptiles and 46 papers (7%) assessed both reptiles and amphibians. In relation to species richness, we identified 1490 amphibian species and 1199 reptile species across all papers; among which 141 taxa were not identified to species level but were still included in our analyses as taxonomic units analogous to species (Online Appendix 2). Among these species, more than half of the studied amphibians were found in South America (537; 38%) and North America (328; 23%), followed by Africa (297; 21%), Asia (137; 10%), Europe (77; 5%), and Oceania (51; 3%). Half of the reptile species studied were from North America (302; 25%) and Africa (278; 23%), with the other half consisting of species from Oceania (276; 23%), South America (200; 17%), Europe (76; 6%), and Asia (67; 6%).
When compared to the known species richness in the world, large portions of European species are studied while species from other continents were severely under-represented (Fig. 2 ). The proportions of amphibian species represented in papers were the highest in Europe (73%), while the proportions are much lower for Africa (23%), North America (23%), South America (18%), Oceania (7%) and Asia (6%) (Fig. 2 a). Among reptiles, Europe represents again the highest proportion of studied species (27%), followed by Oceania (18%), Africa (12%), North America (11%) and South America (8.9%) (Fig. 2 b). In contrast, of all Asian reptile species, only a mere 1.73% were included in the selected papers. The species coverage in our selected papers does not seem optimistic. Amphibians and reptiles each have only six families with more than half of the species covered (including three reptilian families containing one species in total). Meanwhile, 23 and 25 families remain fully neglected for amphibians and reptiles respectively (Figs. 4 – 5 ).
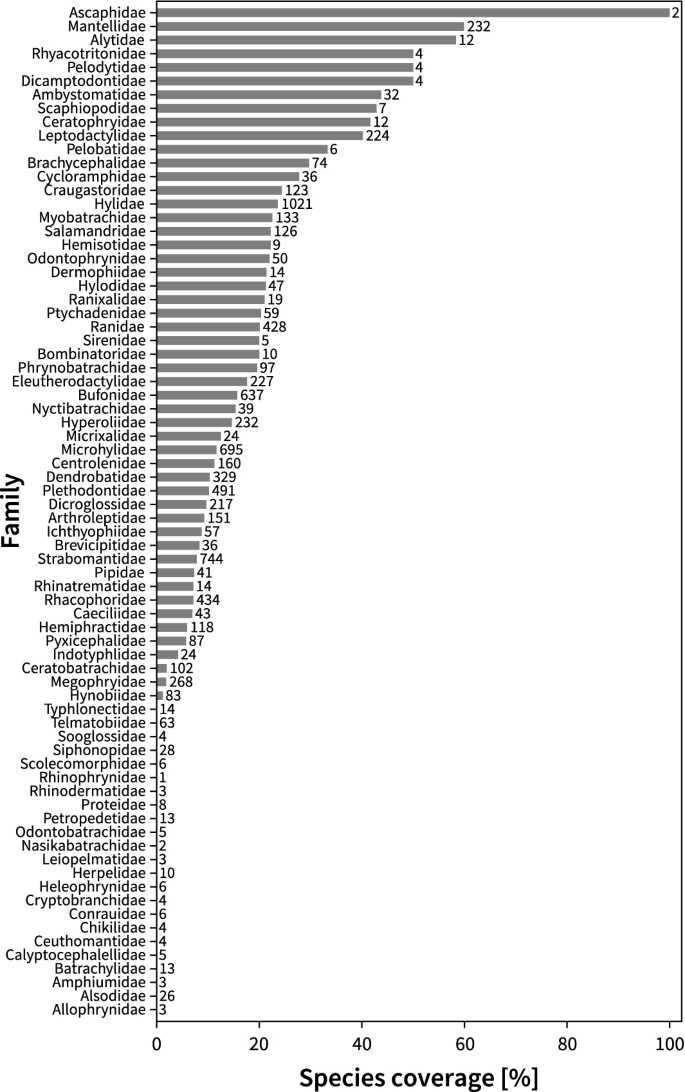
Species coverage for each taxonomic family in selected papers of amphibians. The numbers on each row indicate the total number of species known in its respective family (obtained from AmphibiaWeb 2021 )
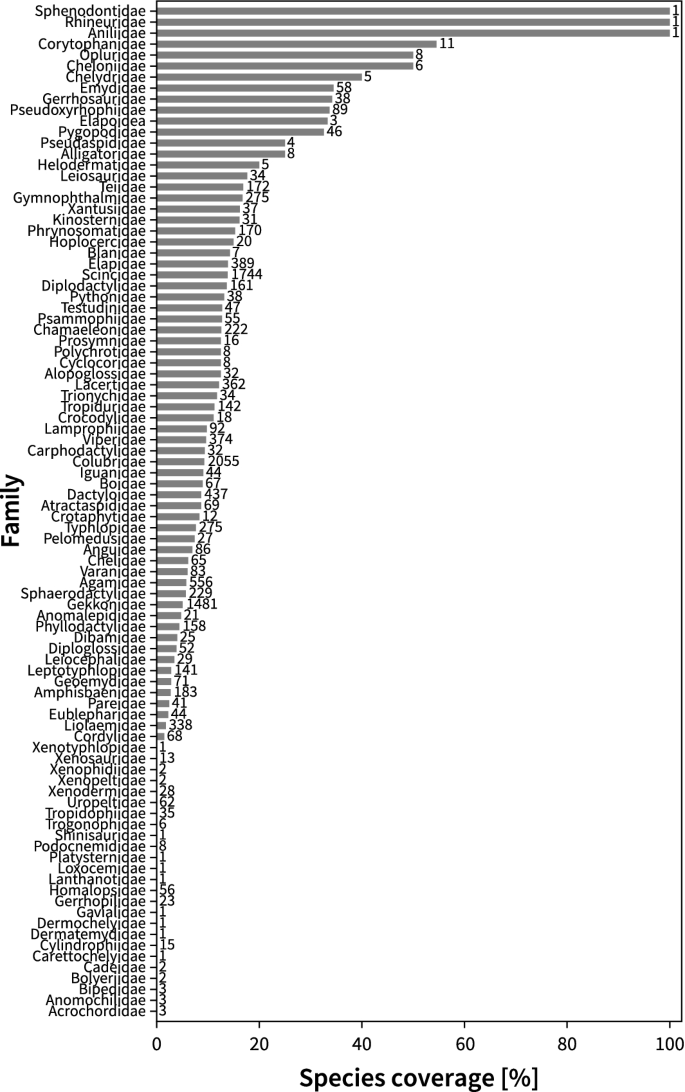
Species coverage for each taxonomic family in selected papers of reptiles. The numbers on each row indicate the total number of species known in its respective family (obtained from ReptileDatabase)
Multiple correspondence analysis provided important insights into underlying patterns in our data allowing us to visualise the relationship between forms of habitat fragmentation (Median = 1 [1–4]), sampling methods (Median = 1 [0–5]) and response variables (Median = 2 [1–6]). Percentage of variance (or eigenvalues) from MCA output represents the contribution of each dimension in explaining the observed patterns. The top ten new dimensions identified by MCA explained a total of 61.64% and 61.16% of the total variance for amphibians and reptiles respectively. The two dimensions with the highest variance percentages explained were found in the first (Dim1, 12.55%) and second (Dim2, 9.13%) dimensions in amphibians (Online Appendix 3–4). Genetics (sampling method; 13.73%) and population (response variable; 12.39%) contributed the most to Dim1, together with species richness (response variable;10.41%) and dispersal (response variable; 9.20%). For Dim2, experimental (sampling method; 14.38%) was the dominant variable, the rest was determined by GIS/Satellite images (sampling method; 9.71%), fitness measure (response variable; 9.12%) and urbanisation (form of fragmentation; 8.94%). For reptiles, the two dimensions explaining the most variation were the first (Dim1, 11.34%) and second (Dim2, 8.28%) dimensions (Online Appendix 3–4). The variables contributing the most to Dim1 were species richness (response variable; 15.51%), abundance (response variable; 10.11%), presence/absence (response variable; 6.97%) and genetics (sampling method; 6.39%). On the other hand, Dim2 was determined by interspecific interaction (response variable; 13.49%), genetics (12.79%), experimental methods (sampling method; 11.21%) and fitness measure (response variable; 10.94%). The contribution of each category to the definition of the dimensions is reported in Online Appendix 3. The categories identified in the MCA dimensions are subsequently used for building the distance matrix in the clustering analysis.
The HCPC analysis identified three clusters of variables for amphibians and reptiles (Online Appendix 5–6). The output of the HCPC analysis is reported in Online Appendix 7. V test represent the influence of variables in the cluster composition. In general, three clusters for both amphibians and reptiles appeared to be uniquely similar by definition of categories (Fig. 6 ). For amphibians, cluster 1 was defined by studies on species richness (p < 0.05, V test = 14.30) and presence/absence (p < 0.05, V test = 13.42), while cluster 2 was determined by experimental studies (p < 0.05, V test = 10.95) and fitness measures (p < 0.05, V test = 9.77). Cluster 3 was defined by genetics (p < 0.05, V test = 18.44) and population studies (p < 0.05, V test = 17.73) (Online Appendix 7). Abundance and functional richness were also unique to cluster 1; other response variables and direct tracking methods were important to cluster 2 and dispersal was present in cluster 3 even though these variables are less expressed (Fig. 6 a).
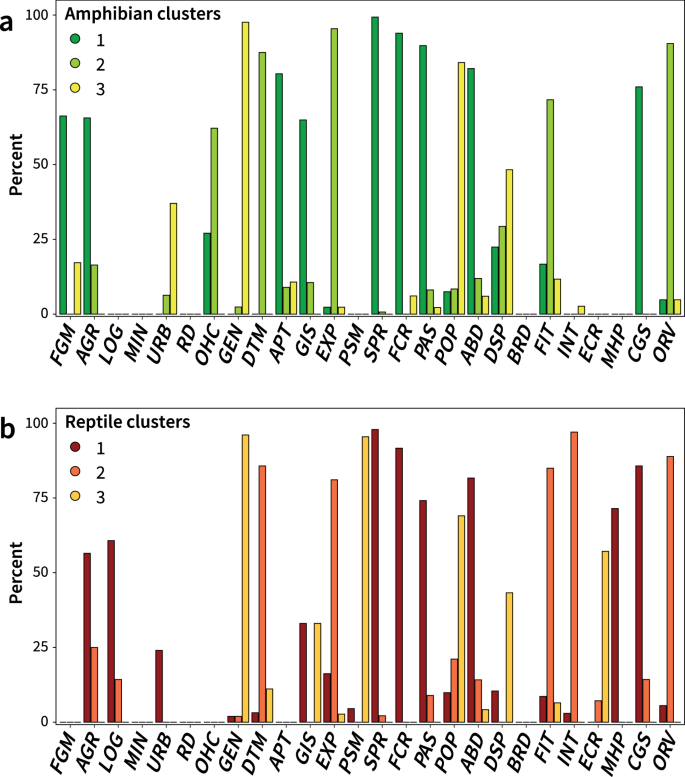
Percentage contribution of the categories contributing to the uniqueness of each cluster in amphibians (Dark green = 1, Bright green = 2, Bright yellow = 3) and reptiles (Dark red = 1, Orange = 2, Dark yellow = 3) based on the Cla/Mod results of HCPC (see Online Appendix 7). Abbreviations for the categories can be found in Fig. 3 and in Online Appendix 1
For reptiles, cluster 1 was represented by species richness (p < 0.05, V test = 14.26), abundance (p < 0.05, V test = 11.22) and presence absence (p < 0.05, V test = 8.55) papers, whereas cluster 2 was determined by papers on fitness measures (p < 0.05, V test = 10.99), direct tracking methods (p < 0.05, V test = 8.64) and interspecific interaction (p < 0.05, V test = 7.86), and cluster 3 was defined by genetics (p < 0.05, V test = 12.79), population (p < 0.05, V test = 9.95) and prediction/simulation models (p < 0.05, V test = 7.68) papers (Online Appendix 7). Although slightly less expressed in the clusters, papers using comparisons between generalist and specialist species and papers on functional richness were also unique to cluster 1; experimental methods and other response variables were heavily present in cluster 2, while dispersal studies were distinct to cluster 3 (Fig. 6 b).
Results from VOSviewer categories of both amphibians and reptiles appear to be similar to each other (Fig. 3 c, d). The clustering of the categories in the co-occurrence network maps confirms what we observed in the HCPC results (Fig. 6 ). In addition to geographical representation of study locations in (1), the corresponding clusters of selected papers are also mapped in Figs. 7 and 8 to investigate the spatial grouping patterns for the three clusters (see Online Appendix 8–9 for geographical representation for each category). We also plotted the temporal trend in Online Appendix 10 and 11. Overall, the three clusters are distributed homogeneously across the globe, but concentrated in the USA, Europe and south eastern Australia. Cluster 1 papers were found to be the most predominant cluster in amphibians (57% papers) across all continents (see Online Appendix 12; Fig. 7 ). When compared to other clusters, studies from this cluster are often conducted in Afrotropics, particularly Madagascar (100% papers), central (Costa Rica (60% papers) and Mexico (92% papers) and south America (80% papers) (Online Appendix 12, Figs. 7 , 8 ). On the other hand, cluster 2 papers appear to be more prevalent for reptile studies compared to amphibian studies, with a higher number of studies conducted across North America (65 to 51) and Australia (22 to 2) (Figs. 7 , 8 ). Lastly, a vast majority of cluster 3 papers were located in North America and Europe (both contributing to 79% of the papers) for amphibians and North America and Australia (both contributing to 84% of the papers) for reptiles (Online Appendix 12, Figs. 7 , 8 ). Publications from this cluster started to gain popularity from 2005 onwards, following similar increasing trends as cluster 2 (Online Appendix 10–11). Overall, except for cluster 1 in South America, most of the clusters in Asia and Africa appear to experience very little or no increase in publications over the years (Online Appendix 10–11).
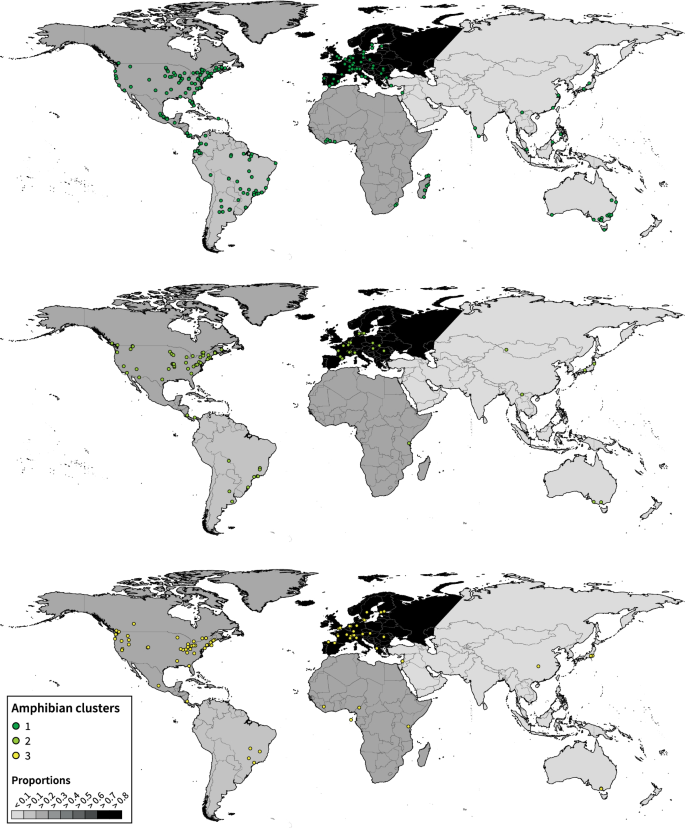
Map of the individual selected papers belonging to each cluster groups (Dark green = 1, Bright green = 2, Bright yellow = 3) for amphibians, with each circle representing the study location. The colour scale of the continents ranging from 0 to 0.9 indicates the proportions of amphibian species represented in the reviewed papers when compared to known species in the world (obtained from AmphibiaWeb)
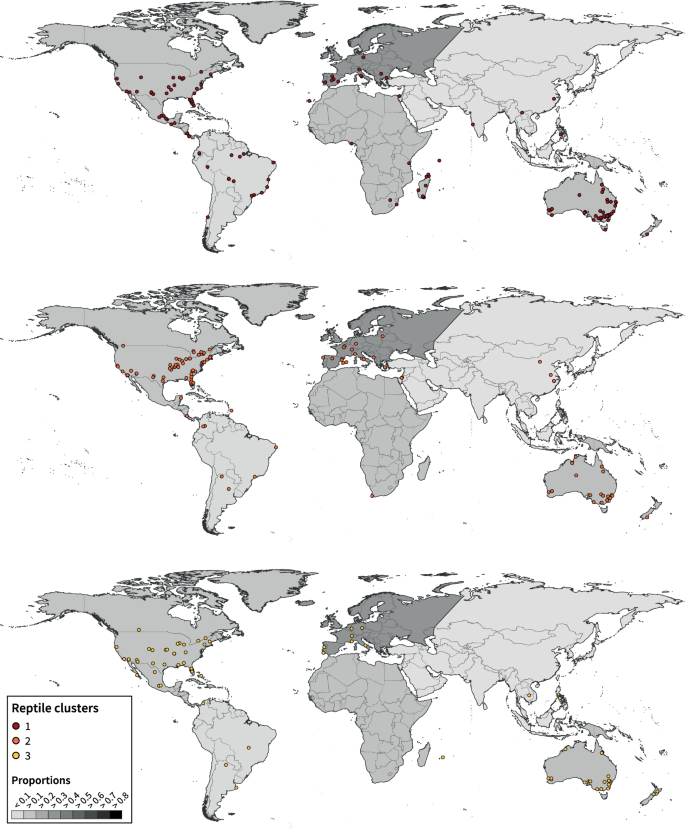
Map of the individual selected papers belonging to each cluster groups (Dark red=1, Orange=2, Dark yellow=3) for reptiles, with each circle representing the study location. The colour scale of the continents ranging from 0.0 – 0.9 indicates the proportions of reptile species represented in the reviewed papers when compared to known species in the world (obtained from ReptileDatabase).
Our review found no improvement in the geographical and taxonomic bias in habitat fragmentation studies for both reptiles and amphibians compared to earlier studies (Fardila et al. 2017 ). Yet, our study has made an effective contribution towards identifying major spatial gaps in habitat fragmentation studies over the past three decades (updating reviews such as Cushman 2006 ; Gardner et al. 2007 )). In particular, we found an overall increase in the number of studies measuring species richness and abundance throughout the years while population-level and genetics studies are still lacking in developing countries. Here, we discuss the issues of (1) biogeographical bias, (2) the extent and focus of habitat fragmentation research and (3) the limitations and knowledge gaps in habitat fragmentation research in herpetology and provide recommendations for future research.
Biogeographical bias
Geographic bias in research papers.
Given the research effort in relatively wealthy countries (Holmgren and Schnitzer 2004 ; Fazey et al. 2005 ) it is not surprising that more than half the papers concern North America and Europe, where there is strong prevalence of herpetological research. This pattern is also evident in other taxonomic groups and biological areas including invasion biology (Pyšek et al. 2008 ), biodiversity conservation (Trimble and Aarde 2012 ; Christie et al. 2020 ), and habitat fragmentation (Fardila et al. 2017 ). The USA alone contributed more than a third of the publications in terms of both authors and location of study (Fazey et al. 2005 ; Melles et al. 2019 ). English speaking countries including the USA, the United Kingdom, and Australia have dominated research output over the last 30 years (Melles et al. 2019 ). These patterns were reflected in the collaboration network maps generated by VOSviewer (Fig. 3 ). Similar hotspots found between who does the research (Fig. 3 ) and the study locations (Fig. 2 ) suggest that authors tend not to move much and only to study ecosystems near to where they are based (Meyer et al. 2015 ). One reason for this bias is the distance to field sites accentuated by the costs and time of travelling.
However, the near absence of studies from many parts of the world that are currently under extreme pressures of habitat loss and degradation are of great concern (Habel et al. 2019 ). We feel that the level of threat associated with habitat fragmentation in these continents is not proportional to the level of research attention required. Naturally biodiverse but less economically developed Southeast Asian and sub-Saharan countries will suffer greatest diversity losses in the next century (Newbold et al. 2015 ). If this persists at the current rate, biodiverse areas will likely disappear before new discoveries in those hotspots are made (Moura and Jetz 2021 ). Although conversely our study found that among other developing countries Brazil is currently conducting relatively more in-country amphibian studies and collaboration with other countries. However, how much of this information reaches decision makers and practitioners remains unknown. This is largely due to the lack of intermediary evidence bridges (Kadykalo et al. 2021 ). These intermediaries identify evidence summaries based on research and priorities and distribute them to practitioners, facilitating exchange of knowledge between and among researchers and practitioners (Holderegger et al. 2019 ; Kadykalo et al. 2021 ).
Geographic bias in focal groups
Congruent to results reported in Gardner et al. ( 2007 ), studies on amphibians were more abundant than studies on reptiles. Over the past years, there has been a strong focus on amphibian population declines. This was catalysed by the emergence of chytridiomycosis and global decline of amphibians (Fisher and Garner 2020 ). Amphibians, and predominantly frogs, are the principal focus of herpetological research, with the highest allocation of resources and the highest publication rates (Ferronato 2019 ). Another reason for this bias may be that amphibians serve as valuable indicators of environmental stress and degradation owing to their aquatic and terrestrial lifestyle and permeable skin (Green 2003 ). These attributes make them extremely sensitive to changes in temperature and precipitation as well as pollution (Sodhi et al. 2008 ). Lizards, also susceptible to temperature changes, however, are characterised by a high degree of endemism, restricted geographic ranges, late maturity, a long life-span and are thus very susceptible to population declines (Todd et al. 2010 ; Meiri et al. 2018 ). Certain groups of reptiles, such as worm lizards and blind snakes lead cryptic and solitary lives in contrast to the large breeding aggregations and choruses of, for example, frogs. Such characteristics make them difficult for researchers to study as they require large amount of search effort for little data (Thompson 2013 ).
- Taxonomic bias
We found a heightened geographical bias in the taxonomic coverage of studies. Given the sheer number of selected papers investigated, it is not surprising that the continents of North and South America cover more than half of the amphibian species studied whereas North America and Africa cover almost half of the reptile species studied. This trend broadly mirrors the geographic distribution pattern of the global described species in both these taxa (AmphibiaWeb 2021 ; Uetz et al. 2021 ). While a large proportion of the known European and North American families such as Alytidae and Ambystomatidae have been investigated (Fig. 4 ), species from other continents remain severely under-represented. Yet, the European continent represents only 2% of the described species globally. This high research intensity bias in low biodiverse regions of the world has been noted previously (Fazey et al. 2005 ). In general, reptiles and amphibians have been disproportionately poorly studied in the tropics and in developing areas despite that these areas show among the highest rates of deforestation and a corresponding rise in the number of threatened species (Böhm et al. 2013 ; Deikumah et al. 2014 ). These biodiverse areas largely consist of threatened species having restricted home ranges (Meiri et al. 2018 ). Even though we observed a great fraction of the species investigated in the Afrotropics (Vallan 2002 ; Hillers et al. 2008 ; Ofori‐Boateng et al. 2013 ; Riemann et al. 2015 ; Blumgart et al. 2017 ), especially Madagascar (see Mantellidae and Opluridae in Fig. 4 ), it seems insufficient when considering that an estimated 3.94 million hectares of forest area of the continent was cleared yearly over the last century (FAO and UNEP 2020 ). Further, biodiverse hotspots such as the neotropical regions and Indo-Malayan tropics have the highest chances of new species of amphibians and reptiles being discovered (Moura and Jetz 2021 ).
Being herpetofauna diversity hotspots, countries in South America and Asia are indeed understudied. Although Brazil has a high number of amphibian studies, less than one percent of known reptile species was studied in both continents (Fig. 2 ). A number of factors contribute to this lack of representation. First, there is an overwhelming number of new species being discovered every year in these hotspots (Moura et al. 2018 ; Moura and Jetz 2021 ). Furthermore, newly discovered species tend to belong to more secretive groups such as burrowing snakes, worm lizards and caecilians (Colli et al. 2016 ). Yet, these fossorial organisms are clearly neglected in fragmentation studies (see Fig. 4 – 5 ) with researchers focusing on well-known taxonomic groups (Böhm et al. 2013 ). On a positive note, despite having the country (Australia) with the highest reptile diversity (Uetz et al. 2021 ), Oceania represented a fair coverage of reptile diversity compared to other continents. Since 2001, there has been an increase of fragmentation studies in Australia (e.g., Brown 2001 ; Mac Nally and Brown 2001 ; Hazell et al. 2001 ) and there is a continuing increase in research output (Melles et al. 2019 ), contributing 85 out of 89 reviewed studies in Oceania over the last 30 years.
Extent and focus of research
Our findings showed important associations between methods and response metrics but not different forms of habitat fragmentation. This either suggests that researchers were not favouring any sampling method and response variable for evaluating the effects of certain habitat fragmentation or this pattern may occur due to a relatively even split of papers dealing with different forms or combinations of habitat fragmentation in the clusters. In general, species richness or diversity appears to explain most of the variation in our data ( see Online Appendix 4 ). While species richness remains a popular diversity metric employed in conservation biology (Online Appendix 12; also see Gardner et al. 2007 ), we also found an increasing trend in the use of genetic techniques for habitat fragmentation studies. More specifically in recent years, molecular genetics have become popular and are often studied together with population connectivity to capture species responses to habitat fragmentation ( see Online Appendix 4 ) (Keyghobadi 2007 ). The HCPC approach identified three main clusters of research fields which will be referred to as research agendas from here onwards. Contrary to our expectation, we did not find a global spatial pattern of research agendas, but instead a rather homogeneous distribution of papers, possibly due to the lack of selected studies which are found in developing countries outside USA, Europe and Australia (Figs. 7 , 8 ). This nevertheless indicates that different sampling methods are shared and used between leading herpetological experts from different countries and that there are continuing collaborations between countries, particularly in North America and Europe.
Below, we describe the research agendas and their corresponding categories (Fig. 6 ) that have contributed significantly to the study of habitat fragmentation for the past 30 years: (a) Agenda 1: Measures of direct individual species responses, (b) Agenda 2: Physiological and movement ecology, and (c) Agenda 3: Technology advancement in conservation research.
Agenda 1: Measures of direct individual species responses
We found that the majority of studies around the globe evaluated patterns of assemblage richness, species presence/absence, and abundance (Figs. 7 , 8 ). These simple patterns of richness, diversity and abundance are the most common responses measured because they provide a good indication of species response to habitat fragmentation and are easy to calculate (Colwell 2009 ). Although species richness does not consider abundance or biomass but treats each species as of equal importance to diversity, species evenness weighs each species by its relative abundance (Hill 1972). Further, composite measures like species diversity indices (e.g., Simpson’s 1/D or Shannon’s H) combine both richness and evenness in a single measure (Colwell 2009 ), preventing biases in results. However, directly measuring these species responses might not be ecologically relevant as they fail to account for patterns in species assemblage turnover. In fact, few selected papers (38 out of 697) in our study have attempted to categorise species into meaningful functional groups or guilds, despite that the categorisation of ecological functions such as habitat preference, taxonomic family, reproductive mode, and body size can be easily done (but see Knutson et al. 1999 ; Peltzer et al. 2006 ; Moreira and Maltchik 2014 ). Knutson et al.( 1999 ) was the first in our selected papers to group species with similar life-history characteristics into guilds and to examine their responses to landscape features. They observed negative associations between urban land use and anuran guilds. Analyses of guilds or functional groups can reveal contradictory results (but not always, see Moreira and Maltchik 2014 ). For example, the species richness of anurans in logged areas of West Africa is found to be as high as in primary habitat (Ernst et al. 2006 ). Yet, analyses of functional groups indicated significantly higher diversity in primary forest communities (Ernst et al. 2006 ). Similar differences were also observed for species with varying degrees of niche overlaps, habitat specialists, and for different continents (Ernst et al. 2006 ; Seshadri 2014 ). These results underline that species richness alone is a poor indicator of the functional value of species in the ecosystem as the relationships between functional diversity and species richness are inconsistent and can sometimes be redundant (functional diversity remains constant if assemblages are functionally similar; Riemann et al. 2017 ; Palmeirim et al. 2017 ; Silva et al. 2022 ). The results of some species richness studies may consequently provide misleading inferences regarding consequences of habitat fragmentation and conservation management (Gardner et al. 2007 ).
Although not substantially greater than the agendas 2 and 3, the measure of individual species responses has always been popular across the globe but also increasingly popular in the tropical and subtropical regions (e.g., South America and Africa; Online Appendix 10–11). For example, a research team led by Mark-Oliver Roedel from Germany has conducted numerous studies on Afrotropical amphibian communities (Hillers et al. 2008 ; Ofori‐Boateng et al. 2013 ; Riemann et al. 2017 ). Due to the higher biodiversity and species rarity in these regions compared to temperate areas, it is reasonable to expect a greater level of sampling effort in patterns of species richness, abundance, and guild assemblage to obtain comparisons of diversity with sufficient statistical power across different land use changes (Gardner et al. 2007 ). Access to highly specific expertise and most up to date methods and technology may not be available in these regions, and as such, study designs are limited to multispecies survey addressing simple patterns of diversity and species assemblages (Hetu et al. 2019 ). Unfortunately at the same time, these forest biomes holding the highest richness and abundance of amphibians and reptiles have showed consistent negative responses to land use changes (Cordier et al. 2021 ).
Agenda 2: physiological and movement ecology
We did not observe a strong association between occupancy and dispersal in our study. Perhaps this is because only a few papers investigated dispersal via habitat occupancy compared to the overwhelming proportions of papers examining the presence of species in response to habitat fragmentation in research agenda 1. Similarly, few studies measure dispersal with direct tracking methods, with the majority that discussed dispersal being based on indirect inferences, such as genetic divergence (see Fig. 3 c, d; Driscoll et al. 2014 ). Genetic approaches can be effective in situations where more direct approaches are not possible (Lowe and Allendorf 2010 ). For instance, using microsatellites and mitochondrial DNA, Buckland et al. ( 2014 ) found no migration occurring between isolated subpopulations of a forest day gecko ( Phelsuma guimbeaui ) in a fragmented forest and predicted a dramatic decrease in survival and allelic diversity in the next 50 years if no migration occurs (Buckland et al. 2014 ). In some cases, molecular markers also allow direct dispersal studies by assigning individuals to their parents or population of origin (Manel et al. 2005 ). However, there are limitations on when these techniques can be applied. Assignment tests require appropriate choices of molecular markers and sampling design to permit quantification of indices of dispersal (Broquet and Petit 2009 ; Lowe and Allendorf 2010 ). Parent–offspring analysis is constrained by the uncertainty in assessing whether offspring dispersal is completed at the time of sampling and sample size (Broquet and Petit 2009 ). Genetic tools may thus be best applied in combination with direct approaches because they contain complementary information (Lowe and Allendorf 2010 ; Safner et al. 2011 ; Smith et al. 2016 ).
Traditional approaches in habitat fragmentation research like radiotracking or capture-mark-recapture of animals can be effective in evaluating dispersal and ecological connectivity between populations. For example, based on mark-recapture data over a nine year period, facultative dispersal rates in an endangered amphibian ( Bombina variegata ) were found to be sex biased and relatively low from resulting patch loss (Cayuela et al. 2018 ). In our case, direct tracking methods are more commonly and effectively used in examining the impacts of habitat modification on changes in ecology directly relating to fitness (Fig. 6 ): home ranges (Price-Rees et al. 2013 ), foraging grounds (MacDonald et al. 2012 ) and survival rates (Breininger et al. 2012 ). Yet, such routine movements associated with resource exploitation do not reflect the biological reality and evolutionary consequences of how organisms change as landscape changes (Van Dyck and Baguette 2005 ). Instead, directed behavioural movements affecting dispersal processes (emigration, displacement or immigration) are crucial in determining the functional connectivity between populations in a fragmented landscape (Bonte et al. 2012 ). In one study, spotted salamanders Ambystoma maculatum tracked with fluorescent powder exhibited strong edge mediated behaviour when dispersing across borders between forest and field habitats and can perceive forest habitats from some distance (Pittman and Semlitsch 2013 ). Knowing such behaviour rules can improve predictions of the effects of habitat configuration on survival and dispersal. However, ongoing conversion of natural ecosystems to human modified land cover increases the need to consider various cover types that may be permeable to animal movements. As such, experimental approaches can be effective in examining the effect of matrix type on species movements as seen in our results (Fig. 6 ) (Rothermel and Semlitsch 2002 ; Mazerolle and Desrochers 2005 ). For example, researchers conducted experimental releases of post-metamorphic individuals of forest amphibians into different substrates and mapped the movements of paths and performance (Cline and Hunter Jr 2016 ). They showed that non-forest matrices with lower structural complexity influence the ability of frogs to travel across open cover and to orient themselves towards the forest from distances greater than 40–55 m. Therefore, it is inaccurate to assume matrix permeability to be uniform across all open-matrix types, particularly in amphibians (Cline and Hunter 2014 , 2016 ).
In addition, the ability to move and disperse is highly dependent on the range of external environments and internal physiological limits (Bonte et al. 2012 ), especially in reptiles and amphibians (Nowakowski et al. 2017 ). The study of physiological effects on movement was seen throughout our selected studies (Fig. 6 ). For example, higher temperatures and lower soil moisture in open habitats could increase evaporative water loss in salamanders (Rothermel and Semlitsch 2002 ). Other tests including interaction effects between landscape configuration and physiological constraints (e.g., dehydration rate Rothermel and Semlitsch 2002 ; Watling and Braga 2015 ); body size (Doherty et al. 2019 ) can be useful to better understand fitness and population persistence. We argue here that multidisciplinary projects examining movement physiology, behaviour and environmental constraints in addition to measuring distance moved are needed to progress this field.
Our results indicate a high bias of agenda 2 papers represented among developed countries, with a strong focus on reptiles compared to amphibians (Price-Rees et al. 2013 ; Doherty et al. 2019 ) (Online Appendix 12, Figs. 7 , 8 ). The adoption of direct tracking as well as genetic methods can be cost prohibitive in developing and poorer regions. However, cheaper and simpler methods to track individuals are increasing (Mennill et al. 2012 ; Cline and Hunter 2014 , 2016 ). Although existing application might not be ideal for reptiles and amphibians, new technologies for tagging and tracking small vertebrates are being developed including acoustic surveys and improved genetic methods (Broquet and Petit 2009 ; Mennill et al. 2012 ; Marques et al. 2013 ). While there are many improvements needed to obtain better quality dispersal data studies on movement ecology, reptiles and amphibians still only account for a mere 2.2% of the studies on dispersal when compared to plants and invertebrates which comprised over half of the studies based on a systematic review (Driscoll et al. 2014 ). Thus, we urge more studies to be conducted on these lesser-known taxa, especially in biodiverse regions. Given the limited dispersal in amphibians and reptiles, having a deeper understanding on their dispersal can be critical for the effective management and conservation of populations and metapopulations (Smith and Green 2005 ).
Agenda 3: technology advancement in conservation research
While community level approaches such as responses in species richness, occupancy, and abundance measure biodiversity response to habitat fragmentation, they are limited in inference because they do not reflect patterns of fitness across environmental gradients and landscape patterns. Instead, genetic structure at the population level can offer a higher resolution of species responses (Manel and Holderegger 2013 ). For instance, genetic erosion heavily affects the rate of species loss in many amphibian species (Allentoft and O’Brien 2010 ; Rivera‐Ortíz et al. 2015 ). Over the past decades we have seen a rapid increase in studies applying genetic analysis to assess the effects of habitat fragmentation (Keyghobadi 2007 ), reflecting the strength of these approaches. This growth is mostly evident in North America and Europe (but also Oceania for reptiles) ( Online Appendix 10–11). The availability of different genetic markers has been increasing, from microsatellites in the 1990s then shifting towards genotyping by sequencing (NGS) technologies that enable rapid genome-wide development (Allendorf et al. 2010 ; Monteiro et al. 2019 ). However, the study of population structure alone can lead to misleading results as environmental changes to species dynamics are not considered. The resistance imposed by landscape features on the dispersal of animals can ultimately shape gene flow and genetic structure (Bani et al. 2015 ; Pilliod et al. 2015 ; Monteiro et al. 2019 ).
To understand this, researchers combine genetic, land cover and climate variables to study the gene flow patterns across heterogeneous and fragmented landscapes (Manel and Holderegger 2013 ). Spatial analyses can be a powerful tool for monitoring biodiversity by quantifying environmental and landscape parameters. The growing interest in both landcover data and the rapid development of computer processing power prompted the development of new prediction methods, primarily in spatial models (Ray et al. 2002 ), ecological niche modelling (Urbina-Cardona and Loyola 2008 ; Tan et al. 2021 ), and landscape connectivity (Cushman et al. 2013 ; Ashrafzadeh et al. 2019 ). In some cases, niche models are useful in assessing the effectiveness of protected areas for endangered species (Urbina-Cardona and Loyola 2008 ; Tan et al. 2021 ).
The integration of genetic data in ecological niche models for recognising possible dispersal movements between populations were observed in our study (Fig. 3 c, d), especially in reptiles (Fig. 6b ). The hallmark of landscape genetics is the ability to estimate functional connectivity among populations and offer empirical approach of adaptive genetic variation in real landscapes to detect environmental factors driving evolutionary adaptation. The most common approach of landscape genetics is determining whether effective distances as determined by the presence of suitable habitat between populations, better predict genetic distances than do Euclidean distances (assuming spatially homogeneous landscape). However, straight-line geographic distance does not normally reflect true patterns of dispersal as landscape barriers or facilitators in a heterogeneous landscape could strongly affect gene flow (Emel and Storfer 2012 ; Fenderson et al. 2020 ). Therefore, in these cases, ecological distances or landscape resistance can often explain a greater deal of genetic variation between fragmented populations (Cushman 2006 ; Bani et al. 2015 ). Using a combination of habitat suitability modelling (e.g., Maxent, Phillips et al. 2017 ), multiple least-cost paths (LCPs) (Adriaensen et al. 2003 ) and the more recent circuit theory analysis (McRae et al. 2008 ) to investigate landscape resistance can be highly effective predicting potential pathways along which dispersal may occur, hence informing conservation management (Emel and Storfer 2012 ; Bani et al. 2015 ; Pilliod et al. 2015 ). To date, landscape genetics has been shown to be particularly useful in studying organisms with complex life histories (Emel and Storfer 2012 ; Shaffer et al. 2015 ). Yet, the applications of landscape genetics have been limited to contemporary patterns using modern genetic data. Few studies have benefitted from the inclusion of temporal genetic data (Fenderson et al. 2020 ). For example, historical DNA samples and heterochronous analyses could allow us to explore how anthropogenic impacts have affected past genetic diversity and population dynamics (Pacioni et al. 2015 ) and identify areas of future suitability of endangered animals in face of climate change (Nogués-Bravo et al. 2016 ). The possibility to investigate migration through spatiotemporal population connectivity can greatly improve the prediction of species responses under future landscape and climate change scenarios (Fenderson et al. 2020 ).
Population genetic and niche modelling studies for both taxa are rarely found in developing regions of the world, especially in Asia and Africa (Figs. 7 , 8 ). Even though conservation priorities are concentrated in these biodiverse regions, invaluable highly specific expertise such as conservation genetics and other contemporary methodologies might not be readily available due to lack of funding and infrastructure (Hetu et al. 2019 ). Thus, we encourage collaborations with the poorer countries initiated by foreign service providers from developed countries. Contrary to expectations, very few studies on conservation genetics were found in China and Japan despite their vast advances in genetic techniques. Fortunately, China has made substantial progress in the last 20 years in understanding human genetic history and interpreting genetic studies of human diseases (Forero et al. 2016 ) as well as biodiversity conservation (Wang et al. 2020 ), yet the same cannot be said for conservation genetics on reptiles and amphibians (Figs. 7 , 8 ), but see Fan et al. ( 2018 ) and Hu et al. ( 2021 ).
Limitations and knowledge gaps
The forms of habitat fragmentation which we categorised may not reflect the ecological impact in the real world as interactions between different habitat fragmentation forms were not accounted for. Although each of these forms of habitat fragmentation possesses serious environmental consequences, their combination could have severe synergistic impacts (Blaustein and Kiesecker 2002 ). For example, a fragmented landscape is not just reduced and isolated, but subject to other anthropogenic disturbances such as hunting, fire, invasive species, and pollution (Laurance and Useche 2009 ; Lazzari et al. 2022 ). Altered climatic conditions and emerging pathogens such as batrachochytrids can also interact with each other, and other threats (Fisher and Garner 2020 ). The use of habitat suitability models based on climatic scenarios, combined with hydrological and urbanisation models, are effective in detecting best to worst case scenarios and local extinctions, as shown for the spotted marsh frog ( Limnodynastes tasmaniensis ) (Wilson et al. 2013 ).
We acknowledge the bias of scientific research introduced from the limitation of search term to English-speaking literature on the geographic distribution of the papers we sampled (Konno et al. 2020 ; Angulo et al. 2021 ). In Latin American journals for example, we found a number of papers published in Spanish, but unfortunately, they did not fit the criteria of our selection (see Online Appendix 2). Conservation studies written in languages other than English are often published in local journals which do not normally go through international peer review.
The homogeneous distribution of the research agendas across geographical regions in our study may be explained by the lack of studies found in South America, Asia and Africa, preventing us to see a potentially dichotomous spatial pattern among the clusters. However, this reflects the current state of research and the challenges faced in less developed countries.
(4) Our study did not investigate whether habitat fragmentation has led to an improved or decreased biotic response. Predicting species response to habitat modification has been reviewed countless times (Rytwinski and Fahrig 2012 ; Driscoll et al. 2014 ; Doherty et al. 2020 ; Newbold et al. 2020 ; Cordier et al. 2021 ). Yet, these reviews often yield little or no general patterns (Doherty et al. 2020 ; Cordier et al. 2021 ). Response variables or traits measured are often found to be poor predictors of the impacts of habitat fragmentation. There are two possible explanations for this discrepancy. First, the strength and direction of the responses differs between species, ecophysiological groups (Rothermel and Semlitsch 2002 ), and phylogenetic or functional groups (Mazerolle and Desrochers 2005 ; Nowakowski et al. 2017 ). Second, responses in animals to different types of disturbance may be specific to the ecosystem where they live. Different biogeographic regions or biomes have different characteristics affecting local species (Lindell et al. 2007 ; Blowes et al. 2019 ; Newbold et al. 2020 ; Cordier et al. 2021 ).
Conclusions and recommendations
Our results underline promising research fields and geographic areas and may serve as a guideline or starting point for future habitat fragmentation studies. We suspect similar paradigms of geographic and thematic patterns to occur in other taxonomic groups.
Although studies dealing with habitat fragmentation impacts on mammals and birds are already widely recognised (Fardila et al. 2017 ), research on reptiles and amphibians has been lacking. We argue that amphibians and reptiles need more attention as they are equally or more threatened but highly neglected (Rytwinski and Fahrig 2012 ; Ferronato 2019 ; Cox et al. 2022 ).
Greater investment is required for studies in tropical and subtropical areas (Segovia et al. 2020 ), especially within the Asian continent. These areas are currently experiencing the highest rates of habitat loss (McDonald et al. 2013 ). Tropical specialists are further restricted to smaller geographic range sizes according to Rapoport’s rule which states that there is a positive latitudinal correlation with range size (Stevens 1989 ) (at least for amphibians in the Northern hemisphere where there is higher temperature and precipitation seasonality; Whitton et al. 2012 ). Having a small range size is often associated with negative responses to habitat modification (Doherty et al. 2020 ). Thus, more effort is needed in developing countries where the crisis is greatest and there is lack of funding and strong language barriers (Fazey et al. 2005 ). There is an urgent need to better integrate studies published in languages other than English with the broader international literature. Useful integration actions include training of local conservation biologists and promoting partnerships and research visits in these regions may have greater conservation consequences to understand global patterns of habitat modification (Meyer et al. 2015 ). Doing so will help remediate the sampling bias towards temperate generalists and will shed light on the fate of tropical specialists.
We encourage improved access to intermediary evidence-based conservation data (Kadykalo et al. 2021 ). Even when well-established genetic and genomic analyses have been proven to be promising area in herpetological conservation (Shaffer et al. 2015 ), there is a general lack of the transfer of knowledge between scientists and practitioners (Holderegger et al. 2019 ). As practitioners are generally interested in species monitoring and the evaluation of success of connectivity measures, an establishment of scientist-practitioner community to facilitate a platform for international exchange would help tremendously in future conservation planning and management (Holderegger et al. 2019 ).
Although different study designs and landscape measures have different strengths and limitations depending on the study objectives, we suggest reporting basic data to describe the effect of habitat fragmentation using standardised sampling methods, indices, and design (Holderegger et al. 2019 ). The results will allow future meta-analyses to be performed.
Incorporate remote sensing data, whenever possible, in studies involving habitat change and fragmentation. The use of niche modelling techniques combined with high resolution remote sensing has been instrumental in detecting potentially fragmented populations. With advances in landscape genomics, we are now able to examine the correlation between environmental factors and genomic data in natural populations (Manel and Holderegger 2013 ; Shaffer et al. 2015 ). Adopting such tools would be valuable in understanding how habitat amounts and configurations affect dispersal, survival, and population dynamics as well as the impacts of anthropogenic changes such as climate change (Shaffer et al. 2015 ).
Data availability
The datasets generated during the current study are available in Online Appendix 1. Codes used in the analyses are available from corresponding author on request.
Adriaensen F, Chardon JP, De Blust G et al (2003) The application of ‘least-cost’ modelling as a functional landscape model. Landsc Urban Plan 64:233–247. https://doi.org/10.1016/S0169-2046(02)00242-6
Article Google Scholar
Allendorf FW, Hohenlohe PA, Luikart G (2010) Genomics and the future of conservation genetics. Nat Rev Genet 11:697–709. https://doi.org/10.1038/nrg2844
Article CAS Google Scholar
Allentoft ME, O’Brien J (2010) Global amphibian declines, loss of genetic diversity and fitness: a review. Diversity 2:47–71. https://doi.org/10.3390/d2010047
AmphibiaWeb (2021) AmphibiaWeb. https://amphibiaweb.org/ . Accessed 22 Feb 2021
Angulo E, Diagne C, Ballesteros-Mejia L et al (2021) Non-English languages enrich scientific knowledge: the example of economic costs of biological invasions. Sci Total Environ 775:144441. https://doi.org/10.1016/j.scitotenv.2020.144441
Ashrafzadeh MR, Naghipour AA, Haidarian M et al (2019) Effects of climate change on habitat and connectivity for populations of a vulnerable, endemic salamander in Iran. Glob Ecol Conserv 19:e00637. https://doi.org/10.1016/j.gecco.2019.e00637
Bani L, Pisa G, Luppi M et al (2015) Ecological connectivity assessment in a strongly structured fire salamander ( Salamandra salamandra ) population. Ecol Evol 5:3472–3485. https://doi.org/10.1002/ece3.1617
Barber PH, Ablan-Lagman MCA, Ambariyanto, et al (2014) Advancing biodiversity research in developing countries: the need for changing paradigms. Bull Mar Sci 90:187–210. https://doi.org/10.5343/bms.2012.1108
Barlow J, França F, Gardner TA et al (2018) The future of hyperdiverse tropical ecosystems. Nature 559:517–526. https://doi.org/10.1038/s41586-018-0301-1
Blaustein AR, Kiesecker JM (2002) Complexity in conservation: lessons from the global decline of amphibian populations. Ecol Lett 5:597–608. https://doi.org/10.1046/j.1461-0248.2002.00352.x
Blowes SA, Supp SR, Antão LH et al (2019) The geography of biodiversity change in marine and terrestrial assemblages. Science 366:339–345. https://doi.org/10.1126/science.aaw1620
Blumgart D, Dolhem J, Raxworthy CJ (2017) Herpetological diversity across intact and modified habitats of Nosy Komba Island, Madagascar. J Nat Hist 51:625–642. https://doi.org/10.1080/00222933.2017.1287312
Böhm M, Collen B, Baillie JEM et al (2013) The conservation status of the world’s reptiles. Biol Conserv 157:372–385. https://doi.org/10.1016/j.biocon.2012.07.015
Bonte D, Van Dyck H, Bullock JM et al (2012) Costs of dispersal. Biol Rev 87:290–312. https://doi.org/10.1111/j.1469-185X.2011.00201.x
Breininger DR, Mazerolle MJ, Bolt MR et al (2012) Habitat fragmentation effects on annual survival of the federally protected eastern indigo snake: indigo snake survival. Anim Conserv 15:361–368. https://doi.org/10.1111/j.1469-1795.2012.00524.x
Broquet T, Petit EJ (2009) Molecular estimation of dispersal for ecology and population genetics. Annu Rev Ecol Evol Syst 40:193–216. https://doi.org/10.1146/annurev.ecolsys.110308.120324
Brown GW (2001) The influence of habitat disturbance on reptiles in a Box-Ironbark eucalypt forest of south-eastern Australia. Biodivers Conserv 10:161–176. https://doi.org/10.1023/A:1008919521638
Buckland S, Cole NC, Groombridge JJ et al (2014) High risks of losing genetic diversity in an endemic Mauritian gecko : implications for conservation. PLoS ONE 9:e93387. https://doi.org/10.1371/journal.pone.0093387
Cayuela H, Besnard A, Quay L et al (2018) Demographic response to patch destruction in a spatially structured amphibian population. J Appl Ecol 55:2204–2215. https://doi.org/10.1111/1365-2664.13198
Christie AP, Amano T, Martin PA et al (2020) The challenge of biased evidence in conservation. Conserv Biol 35:249–262. https://doi.org/10.1111/cobi.13577
Cline BB, Hunter ML Jr (2014) Different open-canopy vegetation types affect matrix permeability for a dispersing forest amphibian. J Appl Ecol 51:319–329. https://doi.org/10.1111/1365-2664.12197
Cline BB, Hunter ML Jr (2016) Movement in the matrix: substrates and distance-to-forest edge affect postmetamorphic movements of a forest amphibian. Ecosphere 7:e01202. https://doi.org/10.1002/ecs2.1202
Colli GR, Fenker J, Tedeschi LG et al (2016) In the depths of obscurity: Knowledge gaps and extinction risk of Brazilian worm lizards (Squamata, Amphisbaenidae). Biol Conserv 204:51–62. https://doi.org/10.1016/j.biocon.2016.07.033
Colwell R (2009) Biodiversity: concepts, patterns, and measurement. The Princeton guide to ecology. Princeton, Princeton University Press, pp 257–263
Chapter Google Scholar
Cordier JM, Aguilar R, Lescano JN et al (2021) A global assessment of amphibian and reptile responses to land-use changes. Biol Conserv 253:108863. https://doi.org/10.1016/j.biocon.2020.108863
Cox N, Young BE, Bowles P et al (2022) A global reptile assessment highlights shared conservation needs of tetrapods. Nature 605:285–290. https://doi.org/10.1038/s41586-022-04664-7
Cushman SA (2006) Effects of habitat loss and fragmentation on amphibians: a review and prospectus. Biol Conserv 128:231–240. https://doi.org/10.1016/j.biocon.2005.09.031
Cushman SA, Shirk AJ, Landguth EL (2013) Landscape genetics and limiting factors. Conserv Genet 14:263–274. https://doi.org/10.1007/s10592-012-0396-0
Deikumah JP, McAlpine CA, Maron M (2014) Biogeographical and taxonomic biases in tropical forest fragmentation research. Conserv Biol J Soc Conserv Biol 28:1522–1531. https://doi.org/10.1111/cobi.12348
Doherty TS, Fist CN, Driscoll DA (2019) Animal movement varies with resource availability, landscape configuration and body size: a conceptual model and empirical example. Landsc Ecol 34:603–614. https://doi.org/10.1007/s10980-019-00795-x
Doherty TS, Balouch S, Bell K et al (2020) Reptile responses to anthropogenic habitat modification: a global meta-analysis. Glob Ecol Biogeogr 29:1265–1279. https://doi.org/10.1111/geb.13091
Driscoll DA, Banks SC, Barton PS et al (2014) The trajectory of dispersal research in conservation biology. Syst Rev PLOS ONE 9:e95053. https://doi.org/10.1371/journal.pone.0095053
Driscoll DA, Armenteras D, Bennett AF et al (2021) How fire interacts with habitat loss and fragmentation. Biol Rev 96:976–998. https://doi.org/10.1111/brv.12687
Emel SL, Storfer A (2012) A decade of amphibian population genetic studies: synthesis and recommendations. Conserv Genet 13:1685–1689. https://doi.org/10.1007/s10592-012-0407-1
Ernst R, Linsenmair KE, Rödel M-O (2006) Diversity erosion beyond the species level: dramatic loss of functional diversity after selective logging in two tropical amphibian communities. Biol Conserv 133:143–155. https://doi.org/10.1016/j.biocon.2006.05.028
Fahrig L (2003) Effects of habitat fragmentation on biodiversity. Annu Rev Ecol Evol Syst 34:487–515. https://doi.org/10.1146/annurev.ecolsys.34.011802.132419
Fahrig L (2017) Ecological responses to habitat fragmentation per se. Annu Rev Ecol Evol Syst 48:1–23. https://doi.org/10.1146/annurev-ecolsys-110316-022612
Fan H, Hu Y, Wu Q et al (2018) Conservation genetics and genomics of threatened vertebrates in China. J Genet Genomics 45:593–601. https://doi.org/10.1016/j.jgg.2018.09.005
FAO and UNEP (2020) The State of the World’s Forests 2020: Forests, biodiversity and people. FAO and UNEP,
Fardila D, Kelly LT, Moore JL, McCarthy MA (2017) A systematic review reveals changes in where and how we have studied habitat loss and fragmentation over 20years. Biol Conserv 212:130–138. https://doi.org/10.1016/j.biocon.2017.04.031
Fazey I, Fischer J, Lindenmayer DB (2005) Who does all the research in conservation biology? Biodivers Conserv 14:917–934. https://doi.org/10.1007/s10531-004-7849-9
Fenderson LE, Kovach AI, Llamas B (2020) Spatiotemporal landscape genetics: investigating ecology and evolution through space and time. Mol Ecol 29:218–246. https://doi.org/10.1111/mec.15315
Ferronato B (2019) An assessment of funding and publication rates in herpetology. Herpetol J. https://doi.org/10.33256/hj29.4.264273
Fisher MC, Garner TWJ (2020) Chytrid fungi and global amphibian declines. Nat Rev Microbiol 18:332–343. https://doi.org/10.1038/s41579-020-0335-x
Forero DA, Wonkam A, Wang W et al (2016) Current needs for human and medical genomics research infrastructure in low and middle income countries. J Med Genet 53:438–440. https://doi.org/10.1136/jmedgenet-2015-103631
Gardner TA, Barlow J, Peres CA (2007) Paradox, presumption and pitfalls in conservation biology: the importance of habitat change for amphibians and reptiles. Biol Conserv 138:166–179. https://doi.org/10.1016/j.biocon.2007.04.017
Gibbons JW, Scott DE, Ryan TJ et al (2000) The Global Decline of Reptiles, Déjà Vu Amphibians: reptile species are declining on a global scale. Six significant threats to reptile populations are habitat loss and degradation, introduced invasive species, environmental pollution, disease, unsustainable use, and global climate change. Bioscience 50:653–666. https://doi.org/10.1641/0006-3568(2000)050[0653:TGDORD]2.0.CO;2
Green DM (2003) The ecology of extinction: population fluctuation and decline in amphibians. Biol Conserv 111:331–343. https://doi.org/10.1016/S0006-3207(02)00302-6
Habel JC, Rasche L, Schneider UA et al (2019) Final countdown for biodiversity hotspots. Conserv Lett 12:e12668. https://doi.org/10.1111/conl.12668
Haddad NM, Brudvig LA, Clobert J et al (2015) Habitat fragmentation and its lasting impact on Earth’s ecosystems. Sci Adv 1:e1500052. https://doi.org/10.1126/sciadv.1500052
Hadley AS, Betts MG (2016) Refocusing habitat fragmentation research using lessons from the last decade. Curr Landsc Ecol Rep 1:55–66. https://doi.org/10.1007/s40823-016-0007-8
Hamer AJ, McDonnell MJ (2008) Amphibian ecology and conservation in the urbanising world: a review. Biol Conserv 141:2432–2449. https://doi.org/10.1016/j.biocon.2008.07.020
Hazell D, Cunnningham R, Lindenmayer D et al (2001) Use of farm dams as frog habitat in an Australian agricultural landscape: factors affecting species richness and distribution. Biol Conserv 102:155–169. https://doi.org/10.1016/S0006-3207(01)00096-9
Hetu M, Koutouki K, Joly Y (2019) Genomics for All: International Open Science Genomics Projects and Capacity Building in the Developing World. Front Genet. https://doi.org/10.3389/fgene.2019.00095
Hillers A, Veith M, Rödel M-O (2008) Effects of forest fragmentation and habitat degradation on West African leaf-litter frogs. Conserv Biol 22:762–772. https://doi.org/10.1111/j.1523-1739.2008.00920.x
Holderegger R, Balkenhol N, Bolliger J et al (2019) Conservation genetics: linking science with practice. Mol Ecol 28:3848–3856. https://doi.org/10.1111/mec.15202
Holmgren M, Schnitzer SA (2004) Science on the rise in developing countries. PLOS Biol 2:e1. https://doi.org/10.1371/journal.pbio.0020001
Hu Y, Fan H, Chen Y et al (2021) Spatial patterns and conservation of genetic and phylogenetic diversity of wildlife in China. Sci Adv 7:eabd5725. https://doi.org/10.1126/sciadv.abd5725
Kadykalo AN, Buxton RT, Morrison P et al (2021) Bridging research and practice in conservation. Conserv Biol 35:1725–1737. https://doi.org/10.1111/cobi.13732
Keyghobadi NK (2007) The genetic implications of habitat fragmentation for animals. Can J Zool. https://doi.org/10.1139/Z07-095
Knutson MG, Sauer JR, Olsen DA et al (1999) Effects of Landscape Composition and Wetland Fragmentation on Frog and Toad Abundance and Species Richness in Iowa and Wisconsin, U.S.A. Conserv Biol 13:1437–1446. https://doi.org/10.1046/j.1523-1739.1999.98445.x
Konno K, Akasaka M, Koshida C et al (2020) Ignoring non-English-language studies may bias ecological meta-analyses. Ecol Evol 10:6373–6384. https://doi.org/10.1002/ece3.6368
Laurance WF, Useche DC (2009) Environmental synergisms and extinctions of tropical species. Conserv Biol 23:1427–1437. https://doi.org/10.1111/j.1523-1739.2009.01336.x
Lazzari J, Sato CF, Driscoll DA (2022) Traits influence reptile responses to fire in a fragmented agricultural landscape. Landsc Ecol 37:2363–2382. https://doi.org/10.1007/s10980-022-01417-9
Lê S, Josse J, Husson F (2008) FactoMineR: an R package for multivariate analysis. J Stat Softw 25:1–18. https://doi.org/10.18637/jss.v025.i01
Lebart L, Morineau A, Piron M (1995) Statistique exploratoire multidimensionnelle. Dunod Paris
Lindell CA, Riffell SK, Kaiser SA et al (2007) Edge responses of tropical and temperate birds. Wilson J Ornithol 119:205–220. https://doi.org/10.1676/05-133.1
Lindenmayer DB, Fischer J (2007) Tackling the habitat fragmentation panchreston. Trends Ecol Evol 22:127–132. https://doi.org/10.1016/j.tree.2006.11.006
Lowe WH, Allendorf FW (2010) What can genetics tell us about population connectivity? Mol Ecol 19:3038–3051. https://doi.org/10.1111/j.1365-294X.2010.04688.x
Mac Nally R, Brown GW (2001) Reptiles and habitat fragmentation in the box-ironbark forests of central Victoria, Australia: predictions, compositional change and faunal nestedness. Oecologia 128:116–125. https://doi.org/10.1007/s004420100632
MacDonald B, Lewison R, Madrak S et al (2012) Home ranges of East Pacific green turtles Chelonia mydas in a highly urbanized temperate foraging ground. Mar Ecol Prog Ser 461:211–221. https://doi.org/10.3354/meps09820
Manel S, Gaggiotti OE, Waples RS (2005) Assignment methods: matching biological questions with appropriate techniques. Trends Ecol Evol 20:136–142. https://doi.org/10.1016/j.tree.2004.12.004
Manel S, Holderegger R (2013) Ten years of landscape genetics. Trends Ecol Evol 28:614–621. https://doi.org/10.1016/j.tree.2013.05.012
Marques TA, Thomas L, Martin SW et al (2013) Estimating animal population density using passive acoustics. Biol Rev 88:287–309. https://doi.org/10.1111/brv.12001
Mazerolle MJ, Desrochers A (2005) Landscape resistance to frog movements. Can J Zool. https://doi.org/10.1139/z05-032
McDonald RI, Marcotullio PJ, Güneralp B (2013) Urbanization and Global Trends in Biodiversity and Ecosystem Services. In: Elmqvist T, Fragkias M, Goodness J et al (eds) Urbanization, Biodiversity and Ecosystem Services: Challenges and Opportunities: A Global Assessment. Springer, Dordrecht, pp 31–52
Google Scholar
McRae BH, Dickson BG, Keitt TH, Shah VB (2008) Using circuit theory to model connectivity in ecology, evolution, and conservation. Ecology 89:2712–2724. https://doi.org/10.1890/07-1861.1
Meiri S, Bauer AM, Allison A et al (2018) Extinct, obscure or imaginary: the lizard species with the smallest ranges. Divers Distrib 24:262–273. https://doi.org/10.1111/ddi.12678
Melles SJ, Scarpone C, Julien A et al (2019) Diversity of practitioners publishing in five leading international journals of applied ecology and conservation biology, 1987–2015 relative to global biodiversity hotspots. Écoscience 26:323–340. https://doi.org/10.1080/11956860.2019.1645565
Mennill DJ, Battiston M, Wilson DR et al (2012) Field test of an affordable, portable, wireless microphone array for spatial monitoring of animal ecology and behaviour. Methods Ecol Evol 3:704–712. https://doi.org/10.1111/j.2041-210X.2012.00209.x
Meyer C, Kreft H, Guralnick R, Jetz W (2015) Global priorities for an effective information basis of biodiversity distributions. Nat Commun 6:8221. https://doi.org/10.1038/ncomms9221
Moher D, Liberati A, Tetzlaff J et al (2009) Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med 6:e1000097. https://doi.org/10.1371/journal.pmed.1000097
Monteiro WP, Veiga JC, Silva AR et al (2019) Everything you always wanted to know about gene flow in tropical landscapes (but were afraid to ask). PeerJ 7:e6446. https://doi.org/10.7717/peerj.6446
Moreira LFB, Maltchik L (2014) Does organic agriculture benefit anuran diversity in rice fields? Wetlands 34:725–733. https://doi.org/10.1007/s13157-014-0537-y
Moura MR, Jetz W (2021) Shortfalls and opportunities in terrestrial vertebrate species discovery. Nat Ecol Evol 5:631–639. https://doi.org/10.1038/s41559-021-01411-5
Moura MR, Costa HC, Peixoto MA et al (2018) Geographical and socioeconomic determinants of species discovery trends in a biodiversity hotspot. Biol Conserv 220:237–244. https://doi.org/10.1016/j.biocon.2018.01.024
Newbold T, Hudson LN, Phillips HRP et al (2014) A global model of the response of tropical and sub-tropical forest biodiversity to anthropogenic pressures. Proc R Soc B Biol Sci 281:20141371. https://doi.org/10.1098/rspb.2014.1371
Newbold T, Hudson LN, Hill SLL et al (2015) Global effects of land use on local terrestrial biodiversity. Nature 520:45–50. https://doi.org/10.1038/nature14324
Newbold T, Oppenheimer P, Etard A, Williams JJ (2020) Tropical and Mediterranean biodiversity is disproportionately sensitive to land-use and climate change. Nat Ecol Evol 4:1630–1638. https://doi.org/10.1038/s41559-020-01303-0
Nogués-Bravo D, Veloz S, Holt BG et al (2016) Amplified plant turnover in response to climate change forecast by Late Quaternary records. Nat Clim Change 6:1115–1119. https://doi.org/10.1038/nclimate3146
Nowakowski AJ, Watling JI, Whitfield SM et al (2017) Tropical amphibians in shifting thermal landscapes under land-use and climate change. Conserv Biol 31:96–105. https://doi.org/10.1111/cobi.12769
Ofori-Boateng C, Oduro W, Hillers A et al (2013) Differences in the Effects of Selective Logging on Amphibian Assemblages in Three West African Forest Types. Biotropica 45:94–101. https://doi.org/10.1111/j.1744-7429.2012.00887.x
Pacioni C, Hunt H, Allentoft ME et al (2015) Genetic diversity loss in a biodiversity hotspot: ancient DNA quantifies genetic decline and former connectivity in a critically endangered marsupial. Mol Ecol 24:5813–5828. https://doi.org/10.1111/mec.13430
Palmeirim AF, Vieira MV, Peres CA (2017) Herpetofaunal responses to anthropogenic forest habitat modification across the neotropics: insights from partitioning β-diversity. Biodivers Conserv 26:2877–2891. https://doi.org/10.1007/s10531-017-1394-9
Peltzer PM, Lajmanovich RC, Attademo AM, Beltzer AH (2006) Diversity of anurans across agricultural ponds in Argentina. In: Hawksworth DL, Bull AT (eds) Marine, Freshwater, and Wetlands Biodiversity Conservation. Springer, Dordrecht, pp 131–145
Phillips SJ, Anderson RP, Dudík M et al (2017) Opening the black box: an open-source release of Maxent. Ecography 40:887–893. https://doi.org/10.1111/ecog.03049
Pilliod DS, Arkle RS, Robertson JM et al (2015) Effects of changing climate on aquatic habitat and connectivity for remnant populations of a wide-ranging frog species in an arid landscape. Ecol Evol 5:3979–3994. https://doi.org/10.1002/ece3.1634
Pittman SE, Semlitsch RD (2013) Habitat type and distance to edge affect movement behavior of juvenile pond-breeding salamanders. J Zool 291:154–162. https://doi.org/10.1111/jzo.12055
Price-Rees SJ, Brown GP, Shine R (2013) Spatial ecology of bluetongue lizards ( Tiliqua spp.) in the Australian wet–dry tropics. Austral Ecol 38:493–503. https://doi.org/10.1111/j.1442-9993.2012.02439.x
Pyšek P, Richardson DM, Pergl J et al (2008) Geographical and taxonomic biases in invasion ecology. Trends Ecol Evol 23:237–244. https://doi.org/10.1016/j.tree.2008.02.002
R Core Team (2021) R: A language and environment for statistical computing
Ray N, Lehmann A, Joly P (2002) Modeling spatial distribution of amphibian populations: a GIS approach based on habitat matrix permeability. Biodivers Conserv 11:2143–2165. https://doi.org/10.1023/A:1021390527698
Riemann JC, Ndriantsoa SH, Raminosoa NR et al (2015) The value of forest fragments for maintaining amphibian diversity in Madagascar. Biol Conserv 191:707–715. https://doi.org/10.1016/j.biocon.2015.08.020
Riemann JC, Ndriantsoa SH, Rödel M-O, Glos J (2017) Functional diversity in a fragmented landscape — Habitat alterations affect functional trait composition of frog assemblages in Madagascar. Glob Ecol Conserv 10:173–183. https://doi.org/10.1016/j.gecco.2017.03.005
Riva F, Fahrig L (2022) Protecting many small patches will maximize biodiversity conservation for most taxa: the SS > SL principle. Preprints
Rivera-Ortíz FA, Aguilar R, Arizmendi MDC et al (2015) Habitat fragmentation and genetic variability of tetrapod populations. Anim Conserv 18:249–258. https://doi.org/10.1111/acv.12165
Rothermel BB, Semlitsch RD (2002) An Experimental investigation of landscape resistance of forest versus old-field habitats to emigrating juvenile amphibians. Conserv Biol 16:1324–1332. https://doi.org/10.1046/j.1523-1739.2002.01085.x
Roux BL, Rouanet H (2004) Geometric Data Analysis: From Correspondence Analysis to Structured Data Analysis. Springer Science, Dordrecht
Rytwinski T, Fahrig L (2012) Do species life history traits explain population responses to roads? A meta-analysis. Biol Conserv 147:87–98. https://doi.org/10.1016/j.biocon.2011.11.023
Safner T, Miaud C, Gaggiotti O et al (2011) Combining demography and genetic analysis to assess the population structure of an amphibian in a human-dominated landscape. Conserv Genet 12:161–173. https://doi.org/10.1007/s10592-010-0129-1
Segovia ALR, Romano D, Armsworth PR (2020) Who studies where? Boosting tropical conservation research where it is most needed. Front Ecol Environ 18:159–166. https://doi.org/10.1002/fee.2146
Seshadri KS (2014) Effects of Historical Selective Logging on Anuran Communities in a Wet Evergreen Forest, South India. Biotropica 46:615–623. https://doi.org/10.1111/btp.12141
Shaffer HB, Gidiş M, McCartney-Melstad E et al (2015) Conservation genetics and genomics of amphibians and reptiles. Annu Rev Anim Biosci 3:113–138. https://doi.org/10.1146/annurev-animal-022114-110920
Silva DJ, Palmeirim AF, Santos-Filho M et al (2022) Habitat Quality, Not Patch Size, Modulates Lizard Responses to Habitat Loss and Fragmentation in the Southwestern Amazon. J Herpetol 56:75–83. https://doi.org/10.1670/20-145
Smith MA, Green DM (2005) Dispersal and the metapopulation paradigm in amphibian ecology and conservation: are all amphibian populations metapopulations? Ecography 28:110–128. https://doi.org/10.1111/j.0906-7590.2005.04042.x
Smith AL, Landguth EL, Bull CM et al (2016) Dispersal responses override density effects on genetic diversity during post-disturbance succession. Proc R Soc B Biol Sci 283:20152934. https://doi.org/10.1098/rspb.2015.2934
Sodhi NS, Koh LP, Brook BW, Ng PKL (2004) Southeast Asian biodiversity: an impending disaster. Trends Ecol Evol 19:654–660. https://doi.org/10.1016/j.tree.2004.09.006
Sodhi NS, Bickford D, Diesmos AC et al (2008) Measuring the meltdown: drivers of global amphibian extinction and decline. PLoS ONE 3:e1636. https://doi.org/10.1371/journal.pone.0001636
Stevens GC (1989) The latitudinal gradient in geographical range: how so many species coexist in the tropics. Am Nat 133:240–256
Stuart SN, Chanson JS, Cox NA et al (2004) Status and trends of amphibian declines and extinctions worldwide. Science 306:1783–1786. https://doi.org/10.1126/science.1103538
Tan WC, Ginal P, Rhodin AGJ et al (2021) A present and future assessment of the effectiveness of existing reserves in preserving three critically endangered freshwater turtles in Southeast Asia and South Asia. Front Biogeogr. https://doi.org/10.21425/F5FBG50928
Thompson W (2013) Sampling Rare or Elusive Species: Concepts, Designs, and Techniques for Estimating Population Parameters. Island Press, Washington
Todd B, Willson J, Gibbons J (2010) The Global Status of Reptiles and Causes of Their Decline. Ecotoxicology of Amphibians and Reptiles. CRC Press, Boca Raton, pp 47–67
Trimble MJ, van Aarde RJ (2012) Geographical and taxonomic biases in research on biodiversity in human-modified landscapes. Ecosphere 3:art119. https://doi.org/10.1890/ES12-00299.1
Uetz P, Freed P, Aguilar R, Hošek J (2021) The Reptile Database. http://www.reptile-database.org/ . Accessed 6 Mar 2021
Urbina-Cardona JN, Loyola RD (2008) Applying niche-based models to predict endangered-hylid potential distributions: are neotropical protected areas effective enough? Trop Conserv Sci 1:417–445. https://doi.org/10.1177/194008290800100408
Vallan D (2002) Effects of anthropogenic environmental changes on amphibian diversity in the rain forests of Eastern Madagascar. J Trop Ecol 18:725–742
Van Dyck H, Baguette M (2005) Dispersal behaviour in fragmented landscapes: routine or special movements? Basic Appl Ecol 6:535–545. https://doi.org/10.1016/j.baae.2005.03.005
van Eck NJ, Waltman L (2014) Visualizing Bibliometric Networks. In: Ding Y, Rousseau R, Wolfram D (eds) Measuring Scholarly Impact: Methods and Practice. Springer International Publishing, Cham, pp 285–320
Wang W, Feng C, Liu F, Li J (2020) Biodiversity conservation in China: a review of recent studies and practices. Environ Sci Ecotechnology 2:100025. https://doi.org/10.1016/j.ese.2020.100025
Ward JH (1963) Hierarchical grouping to optimize an objective function. J Am Stat Assoc 58:236–244. https://doi.org/10.1080/01621459.1963.10500845
Watling JI, Braga L (2015) Desiccation resistance explains amphibian distributions in a fragmented tropical forest landscape. Landsc Ecol 30:1449–1459. https://doi.org/10.1007/s10980-015-0198-0
Whitton FJS, Purvis A, Orme CDL, Olalla-Tárraga MÁ (2012) Understanding global patterns in amphibian geographic range size: does Rapoport rule? Glob Ecol Biogeogr 21:179–190. https://doi.org/10.1111/j.1466-8238.2011.00660.x
Wilson JN, Bekessy S, Parris KM et al (2013) Impacts of climate change and urban development on the spotted marsh frog ( Limnodynastes tasmaniensis ). Austral Ecol 38:11–22. https://doi.org/10.1111/j.1442-9993.2012.02365.x
Download references
Acknowledgements
W.C. Tan was supported financially through a scholarship by the German Academic Exchange Service (DAAD). This work would not be possible without M. Flecks for his invaluable technical assistance with the figures.
Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and affiliations.
Herpetology Section, LIB, Museum Koenig, Bonn, Leibniz Institute for the Analysis of Biodiversity Change, Adenauerallee 127, 53113, Bonn, Germany
W. C. Tan & D. Rödder
UMR 7179 C.N.R.S/M.N.H.N., Département Adaptations du Vivant, Bâtiment d’Anatomie Comparée, 55 Rue Buffon, 75005, Paris, France
Department of Biology, Evolutionary Morphology of Vertebrates, Ghent University, K.L. Ledeganckstraat 35, 9000, Gent, Belgium
Department of Biology, University of Antwerp, Universiteitsplein 1, B-2610, Antwerpen, Belgium
You can also search for this author in PubMed Google Scholar
Contributions
WCT, AH, and DR contributed to the study idea and conception. Literature search and data collection were performed by WCT and data analysis by DR and WCT. The first draft of the manuscript was written by WCT and all authors critically revised on later versions. All authors read and approved the final manuscript.
Corresponding author
Correspondence to W. C. Tan .
Ethics declarations
Competing interests.
The authors declare no conflicts of interest.
Additional information
Communicated by Ricardo Correia.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Online appendices
Below is the link to the electronic supplementary material.
Captions for appendices (PDF 288 kb)
Appendix 1(pdf 138 kb), appendix 2 (csv 3608 kb), appendix 3 (xlsx 47 kb), appendix 4 (pdf 113 kb), appendix 5 (pdf 293 kb), appendix 6 (pdf 80 kb), appendix 7 (xlsx 18 kb), appendix 8 (pdf 55343 kb), appendix 9 (pdf 55290 kb), appendix 10 (eps 5675 kb), appendix 11 (eps 5665 kb), appendix 12 (xlsx 13 kb), rights and permissions.
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/ .
Reprints and permissions
About this article
Tan, W.C., Herrel, A. & Rödder, D. A global analysis of habitat fragmentation research in reptiles and amphibians: what have we done so far?. Biodivers Conserv 32 , 439–468 (2023). https://doi.org/10.1007/s10531-022-02530-6
Download citation
Received : 18 August 2022
Revised : 02 December 2022
Accepted : 09 December 2022
Published : 08 January 2023
Issue Date : February 2023
DOI : https://doi.org/10.1007/s10531-022-02530-6
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Habitat change
- Herpetofauna
- Geographical bias
- Research agendas
- Systematic review
- Find a journal
- Publish with us
- Track your research
- Open access
- Published: 03 April 2024
NODAL variants are associated with a continuum of laterality defects from simple D-transposition of the great arteries to heterotaxy
- Zain Dardas ORCID: orcid.org/0000-0003-2387-3122 1 na1 ,
- Jawid M. Fatih 1 na1 ,
- Angad Jolly 1 na1 ,
- Moez Dawood 1 , 2 , 3 ,
- Haowei Du 1 ,
- Christopher M. Grochowski 1 ,
- Edward G. Jones 4 ,
- Shalini N. Jhangiani 1 , 2 ,
- Xander H. T. Wehrens 4 , 5 , 6 , 7 ,
- Pengfei Liu 1 , 8 ,
- Weimin Bi 1 , 8 ,
- Eric Boerwinkle 2 , 9 ,
- Jennifer E. Posey 1 ,
- Donna M. Muzny 1 , 2 ,
- Richard A. Gibbs 1 , 2 ,
- James R. Lupski ORCID: orcid.org/0000-0001-9907-9246 1 , 10 , 11 , 2 ,
- Zeynep Coban-Akdemir ORCID: orcid.org/0000-0001-9928-9032 1 , 9 &
- Shaine A. Morris ORCID: orcid.org/0000-0002-8056-0934 4
Genome Medicine volume 16 , Article number: 53 ( 2024 ) Cite this article
182 Accesses
6 Altmetric
Metrics details
NODAL signaling plays a critical role in embryonic patterning and heart development in vertebrates. Genetic variants resulting in perturbations of the TGF-β/NODAL signaling pathway have reproducibly been shown to cause laterality defects in humans. To further explore this association and improve genetic diagnosis, the study aims to identify and characterize a broader range of NODAL variants in a large number of individuals with laterality defects.
We re-analyzed a cohort of 321 proband-only exomes of individuals with clinically diagnosed laterality congenital heart disease (CHD) using family-based, rare variant genomic analyses. To this cohort we added 12 affected subjects with known NODAL variants and CHD from institutional research and clinical cohorts to investigate an allelic series. For those with candidate contributory variants, variant allele confirmation and segregation analysis were studied by Sanger sequencing in available family members. Array comparative genomic hybridization and droplet digital PCR were utilized for copy number variants (CNV) validation and characterization. We performed Human Phenotype Ontology (HPO)-based quantitative phenotypic analyses to dissect allele-specific phenotypic differences.
Missense, nonsense, splice site, indels, and/or structural variants of NODAL were identified as potential causes of heterotaxy and other laterality defects in 33 CHD cases. We describe a recurrent complex indel variant for which the nucleic acid secondary structure predictions implicate secondary structure mutagenesis as a possible mechanism for formation. We identified two CNV deletion alleles spanning NODAL in two unrelated CHD cases. Furthermore, 17 CHD individuals were found (16/17 with known Hispanic ancestry) to have the c.778G > A:p.G260R NODAL missense variant which we propose reclassification from variant of uncertain significance (VUS) to likely pathogenic. Quantitative HPO-based analyses of the observed clinical phenotype for all cases with p.G260R variation, including heterozygous, homozygous, and compound heterozygous cases, reveal clustering of individuals with biallelic variation. This finding provides evidence for a genotypic-phenotypic correlation and an allele-specific gene dosage model.
Our data further support a role for rare deleterious variants in NODAL as a cause for sporadic human laterality defects, expand the repertoire of observed anatomical complexity of potential cardiovascular anomalies, and implicate an allele specific gene dosage model.
The cardiovascular system is among the first physiological functional systems to develop in the vertebrate embryo. Heart development initiates with the formation of the primitive heart tube following the torsional folding of the embryo during the end of the third week of gestation. Once formed, the primitive heart tube must break the pre-existing left-right (L-R) symmetry and undergo a series of septation events that culminate in the formation of a four-chambered heart [ 1 ]. Subtle deviations in heart development can result in congenital heart disease (CHD), which is a collection of defects that together comprise the most prevalent form of birth defect with a birth prevalence of 0.8% of all newborns [ 2 , 3 ]. The etiology of CHD is incompletely understood and certainly due to multiple mechanisms.
One large class of CHD are those related to laterality defects. Classically the laterality defect classification has included situs inversus totalis (complete mirror-image reversal of the chest and abdominal organs usual positions) and heterotaxy (a state of partial rearrangement or anatomical positioning with regard to the body axes) [ 4 ]. However, both animal and human published studies suggest that other CHD lesions could be due to altered laterality development of the heart [ 4 , 5 , 6 , 7 ]. In fact, 3–7% of all apparently isolated CHDs, comprising double outlet right ventricle (DORV), atrioventricular canal defect (AVCD), or transposition of the great arteries (TGA), have been suggested to arise from abnormal embryonic L-R axis patterning [ 5 , 6 ].
The genetics underlying the etiology of laterality defects is heterogeneous and our understanding of the genes involved is limited, but autosomal dominant, autosomal recessive, and X-linked inheritance patterns have each been observed for rare disease traits involving heterotaxy [ 8 , 9 ]. Nevertheless, most clinical cases observed are sporadic in nature and could have de novo mutation contributing or perhaps novel compound inheritance including combinations of biallelic variant alleles contributed from each parent. Over 100 genes, including NODAL , ACVR2B , GDF1 , ZIC3 , SHROOM3 , LZTFL1 , and ciliary genes like DNAH11 , DNAAF1 , and ODAD1 , have been implicated in laterality defects [ 10 ].
A key player in the molecular control of L-R axis development is the NODAL signaling pathway [ 11 ]. Studies in mice revealed that during gastrulation, Nodal, a growth factor from the TGF-ß family, is asymmetrically expressed in the primitive node. Expression is expanded and amplified in the left-lateral plate mesoderm (L-LPM) but inhibited in the right-lateral plate mesoderm (R-LPM) [ 12 ].
Here, we analyzed a large cohort of individuals with clinically diagnosed laterality defects. We found evidence for missense, nonsense, splice site, indels, and/or structural variants in NODAL as potential causes of heterotaxy and other laterality defects in 33 cases. Furthermore, we reinvestigated ClinVar classification of NODAL missense variant NM_018055.5: c.778G > A:p.G260R (Conflicting Interpretation of Pathogenicity into Likely Pathogenic), which was identified in 17/33 cases, and report quantitative phenotypic comparisons of patients with G260R in heterozygous versus biallelic (homozygous and compound heterozygous) states, which implicate a gene dosage effect.
Case ascertainment
Cases were ascertained from 2 sources (Additional file 1 : Tables S1-S3) of subjects with laterality CHD, defined as heterotaxy or congenital heart defects thought to arise by atrio-ventricular discordance or by ventriculo-arterial discordance. Group 1 is a large prospective study of laterality CHD ( n = 583) for which probands were recruited with informed consent to undergo genetic testing based at Baylor College of Medicine (IRB approval number: H-1843). Within this study, 321 of the subjects underwent proband-only exome sequencing (ES) by the Center for Mendelian Genomics (Group 1a). Second (Group 1b), available relevant family members of those found to have a NODAL variant in Group 1a underwent targeted testing. Third, a smaller number of subjects ( n = 269, Group 1c) within Group 1 with some but not complete overlap of Group 1a had previously undergone single-gene sequencing for NODAL variants [ 13 ]. Group 2 is comprised of probands and family members from Texas Children’s Hospital Heart Center with laterality CHD in which the proband underwent microarray or exome or panel sequencing (including NODAL ) which demonstrated pathogenic/likely pathogenic (P/LP) NODAL variants and had CHD were included. For one proband in Group 2, a CNV spanning NODAL was discovered upon reanalysis using the clinical microarray (CMA) data available at Baylor Genetics (BG) (CVG0007). The Institutional Review Board of Baylor College of Medicine approved all research study protocols. Written informed consent was obtained from all participating individuals including probands and any available family members from Group 1. For the clinically tested patients (Group 2), a waiver of consent was granted as part of 2 retrospective cohort studies of clinical genetic testing in CHD (IRB approval numbers: H-48014 and H-41191). Clinical data were ascertained by individual and familial history, as well as review of the medical records. Cardiac phenotypic data were obtained by the review of echocardiograms, magnetic resonance imaging, computed tomography imaging, angiography, and operative reports.
Exome sequencing
ES was performed on genomic DNA for probands from Group 1a at the Human Genome Sequencing Center at Baylor College of Medicine through the Baylor-Hopkins Center for Mendelian Genomics initiative using the Illumina HiSeq 2000 platform and the Mercury pipeline as described previously [ 14 , 15 ]. The methods used for Group 1c ( n = 269) are described in Mohapatra et al. [ 13 ]. For those NODAL variant subjects with CHD ascertained through clinical sequencing, all 5 underwent CMA. Regarding NODAL sequencing, two of the clinical cases underwent trio exome sequencing (CVG0005-fetal and CVG0006), one underwent proband-only sequencing due to unavailability of parental samples (CVG0001), one underwent a trio laterality panel (CVG0003), and one underwent a proband-only laterality panel (CVG0007).
Variant filtering and validation
To detect potential disease-causing NODAL SNVs and indels in exome and panel sequencing, a stepwise analysis workflow was implemented. We investigated homozygous, heterozygous, and compound-heterozygous variant alleles from a 329 gene list (Additional file 1 : Table S4) that includes either known or candidate CHD genes. These genes were collated by combining the genes in the CHD gene database [ 16 ], genes listed on the cardiology panels of Baylor Genetics, Invitae, and Ambry, as well as including known human CHD genes and CHD genes involved in mouse experiments. Rare variants (< 0.01%) were prioritized according to frequency in the population databases including the 1000 Genomes Project (TGP); the Atherosclerosis Risk in Communities Study Database (ARIC); gnomAD; and our in-house-generated exome database (personal genome exomes from ∼ 13,000 individuals) at the BCM-HGSC. Rare variants with a Combined Annotation Dependent Depletion (CADD)-phred score of > 15 were included. Candidate SNVs that remained after the ES analysis process were orthogonally validated and segregated in available family members via an orthogonal approach (Sanger- Dideoxy sequencing).
DNA cloning
To validate the indel variant (p.R234_P241delinsLTS) in families (6 and 7), we performed cloning experiments using the TA Cloning™ Kit (Catalog number: K202020) from Invitrogen™. Primers were designed to amplify a target region (525 bp) flanking the variant. A ligation reaction between pCR2.1 and the target gene was set up with a 1:1 vector/insert molar ratio. The ligation reaction was carried out in 5X T4 DNA ligase reaction buffer at 25 °C for 1 h. Two microliters of the ligation reaction was used to transform 50 μl of One Shot™ TOP10 Chemically Competent E. coli cells (Catalog number: C404003). The transformed cells were single colony purified on Luria-Bertani (LB) plates containing kanamycin and incubated at 37 °C for 16 h. Twelve colonies were randomly selected, inoculated into LB broth, and cultured overnight. Sanger dideoxy sequencing was performed on the cultured colonies.
NODAL CNV analysis
To identify potential CNV deletions spanning NODAL from exome data, we used XHMM (eXome-Hidden Markov Model) [ 17 ], a publicly available bioinformatics tool, and an in-house-developed software, HMZDelFinder [ 18 ]. Furthermore, CNVs spanning NODAL were assessed using the CMA data available at BG.
Human Phenotype Ontology (HPO)-based quantitative phenotypic similarity analysis
A detailed description of the methods used for HPO-based quantitative phenotypic similarity analysis has been previously published [ 19 ]. Briefly, proband cardiac and laterality phenotypes were annotated using HPO terms (Additional file 1 : Table S5). A symmetric Lin similarity score was calculated with the OntologyX suite of R packages [ 20 ] and used to generate pairwise phenotypic similarity scores between all NODAL probands. This similarity matrix was then used to generate a distance matrix for clustering analysis. A gap statistic was calculated for number of clusters 1–15 and plotted to generate a gap statistic curve. The slope of the curve, namely the point at which the slope of the curve had the greatest decrease (4 clusters) was used as a guide for the number of clusters to group probands into. Hierarchical Agglomerative Clustering (HAC) using the Ward method was then used to cluster probands. A heatmap was generated with the ComplexHeatmap [ 21 ] R package based on the previously generated similarity matrix and ordered according to HAC clustering.
Orthogonal validation of predicted NODAL CNV deletions
Droplet digital PCR (ddPCR) and/or array comparative genomic hybridization (aCGH) were performed for variant copy number validation and segregation analysis of two potential NODAL CNV deletions in two unrelated families. ddPCR experiments were performed using the QX200 AutoDG Droplet Digital PCR System according to the manufacturer’s protocols and previously described methods [ 22 ] (Additional file 2 : Table S6 for primer information).
For aCGH, we used an Agilent custom-designed high-resolution array targeting Chr10q (AMADID: 086730). Microarray protocols, including DNA digestion, probe labeling, gender-matched hybridization, and post-washing, were performed as described previously with minor modifications [ 23 ]. Agilent SureScan and Feature Extraction software were utilized to achieve the image-to-digital transition, with further data analysis and visualization on the Agilent Genomic Workbench. Genomic coordinates were described in reference to GRCh37/hg19 assembly.
Breakpoint junction analysis of NODAL CNV deletions
Breakpoint junctions identified in the aCGH data were located and visualized using the Agilent Genomic Workbench. Inward facing primers were designed outside of the deleted regions. Breakpoint junctions were obtained through long-range PCR (LR-PCR) using TaKaRa LA Taq according to the manufacturer’s protocol (TaKaRa Bio Company, Cat.No.RR002). Purified PCR products were sequenced by Sanger dideoxy sequencing (BCM Sequencing Core, Houston, TX, USA). To map the nucleotide-level positions of the breakpoint junctions, the DNA sequences resulting from Sanger sequencing were aligned to the reference genome sequence (UCSC genome browser, GRCh37/hg19).
Genotypic and phenotypic expansion of NODAL SNVs/INDEL in laterality defects
The majority of cases were collected from a prospective study of congenital cardiac laterality defects. Proband exome sequencing was available in 321 of these subjects. While a NODAL SNV or indel had been already reported in 10 of these subjects by our group [ 13 , 15 ], re-analysis detected an additional 11 cases, for a total of 21/321 (6.5%) cases with a NODAL variant and laterality CHD. An additional 6 cases with a NODAL variant were included that were previously reported by our group using single-gene sequencing for NODAL of probands from a separate subcohort of laterality CHD [ 13 ]. For these 27 probands from Group 1 and the 4 probands from Group 2 who had incomplete familial testing, we performed Sanger dideoxy sequencing for variant allele confirmation and segregation analysis in available family members. The NODAL variant was inherited in 19 (including 3 homozygous and 1 compound heterozygous inheritance), de novo in 2, and inheritance data unknown in 10 probands because of lacking information from one or both parents.
Analysis of relatives also revealed an unreported case of an affected aunt (LAT0045, Family 2) with laterality CHD and a NODAL variant (Fig. 1 ). In Group 2, one infant subject that underwent clinical testing (CVG0005) was a son of a proband (LAT0080) in the larger 321-person (Group 1a). These combined cohorts and parental evaluation resulted in 31 unrelated families with NODAL variants; 33 subjects with CHD (Additional file 1 : Tables S1 and S2).
Comprehensive pedigrees and their genotypes for families with NODAL variants. Standard pedigree structures are utilized—filled circles and squares denote clinically affected individuals, and probands are indicated by black arrows. A Pedigrees of probands harboring heterozygous NODAL variants. B Pedigrees of probands harboring biallelic NODAL variants
A total of eleven NODAL SNVs (8 missense, 2 splice site, and 1 nonsense) were detected in 31 laterality CHD cases from 29 families (Table 1 , Fig. 1 ). These variant alleles mainly were localized to the portions of the protein constituting the TGF-β mature domain (Fig. 2 ). Cases with NODAL variants showed various CHD lesions, all of which had abnormal ventricular looping and/or abnormal great artery relationship (Table 2 —summary, Additional file 1 : Table S3-detailed). Within the laterality cohort, NODAL variants were most frequently observed in cases with congenitally corrected transposition of the great arteries (CCTGA), also known as levo-transposition of the great arteries (LTGA) (15.4%) and least frequently in cases with simple dextro-transposition of the great arteries (DTGA) (6.1%) and double outlet right ventricle with malposition of the great arteries (0.0%) (Table 3 ).
Schematic diagram of NODAL protein structure from conceptual translation of transcript NM_018055 with mapping location (vertical lollipops) of the exact map position of amino acid variants observed in this study. In total, we identified twelve different SNV alleles and indel that are distributed among the NODAL domains. The yellow horizontal rectangle represents the TGF-beta propeptide (amino acid 29-166) and the orange horizontal rectangle represents the mature transforming growth TGF-beta domain (amino acid 247-347). Blue circles denote missense variants, green circles denote splice site variants, red circles denote nonsense variants, and brown circles denote a complex indel variant. Numbers inside the circles refer to the variant frequency in our cohort
Two of the identified NODAL missense variants (c.2T>C, p.M1T; c.1039C>T, p.L347F) represented unreported variant alleles that have not been previously associated with laterality defects. The heterozygous p.M1T, identified in proband (LAT0080), was transmitted to his affected son (CVG005). This variant was associated with intrafamilial variable phenotypic expressivity in the proband and his son. The proband LAT0080 presented with heterotaxy, asplenia, right atrial isomerism, mitral atresia, ventricular septal defect (VSD), DORV with D-malposed great arteries (D-MGA), and pulmonary stenosis (PS). He also developed severe arteriovenous malformations (AVMs) and vesicoureteral reflux (VUR); the latter for which he underwent post ureteral reimplantation surgery. Whereas his son (CVG005) exhibited CCTGA with L-ventricular looping, LTGA, pulmonary atresia, and VSD (Table 2 and Fig. 3 ).
Heart depictions illustrating anatomy, blood flow (yellow arrows) and oxygenation (color coded red for oxygenated, blue for unoxygenated, and purple for poorly oxygenated) for different heart defects compared to A the normal heart anatomy. These include B dextro-transposition of the great arteries (DTGA), C double outlet right ventricle with D-malposed great arteries, D tricuspid atresia with D-malposed great arteries, E congenitally corrected transposition of the great arteries (CCTGA) with left ventricular looping and L-transposition of the great arteries, F double inlet left ventricle (DILV) with L-looped ventricles. CCTGA and DILV are illustrated with dextrocardia, but may be levocardic or dextrocardic. G Heterotaxy, asplenia syndrome/right atrial isomerism type
The case LAT0048, harboring a previously unreported p.L347F variant, presented with DTGA. Parental testing revealed that this rare variant was inherited from his father without CHD (Fig. 1 ). Leucine 347 is the last amino acid in the NODAL protein, and its substitution to phenylalanine is predicted to have a deleterious effect on NODAL function with a CADD score of 31 and a REVEL score of 0.51.
A recurrent indel allele potentially caused by secondary structure mutagenesis
We identified one indel variant (p.R234_P241delinsLTS, c.700_723delins) in two unrelated cases (LAT0191 and LAT1763; families 6 and 7, respectively) (Table 1 and Fig. 4 A, B). To facilitate variant interpretation and validation, we performed cloning experiments, to enable separation of alleles, on the probands and parental DNA for subsequent Sanger sequencing. These experiments confirmed that these two probands did not have frameshift variants, but indeed have the nonframeshift delinsLTS variant allele. Of note, the secondary structure predictions for “wild-type” (WT) and mutant single-strand nucleic acid using the RNAfold Server available at http://rna.tbi.univie.ac.at/ , applying the minimum free energy (Fig. 4 C) and thermodynamic ensemble (Fig. 4 D) functions for intramolecular W-C base pairing, suggest that this complex mutation is mediated by secondary structure mutagenesis. The WT structure has a minimum free energy of −10.40 kcal/mol, while the mutant structure has a minimum free energy of −13.20 kcal/mol (lower by 2.80 kcal/mol). For Family 7 (LAT1763), in addition to the c.700_723delins variant allele, we also detected the c.778G > A:p.G260R variant. Allele 1 in LAT1763 is shown to be WT for the c.700_723 locus, while containing the c.778G > A variant. Allele 2 was shown to contain the c.700_723delins variant while being WT at the c.778G > A locus, confirming these variant alleles are in a trans configuration and thus represent a compound heterozygous combination of biallelic variation (Fig. 4 ).
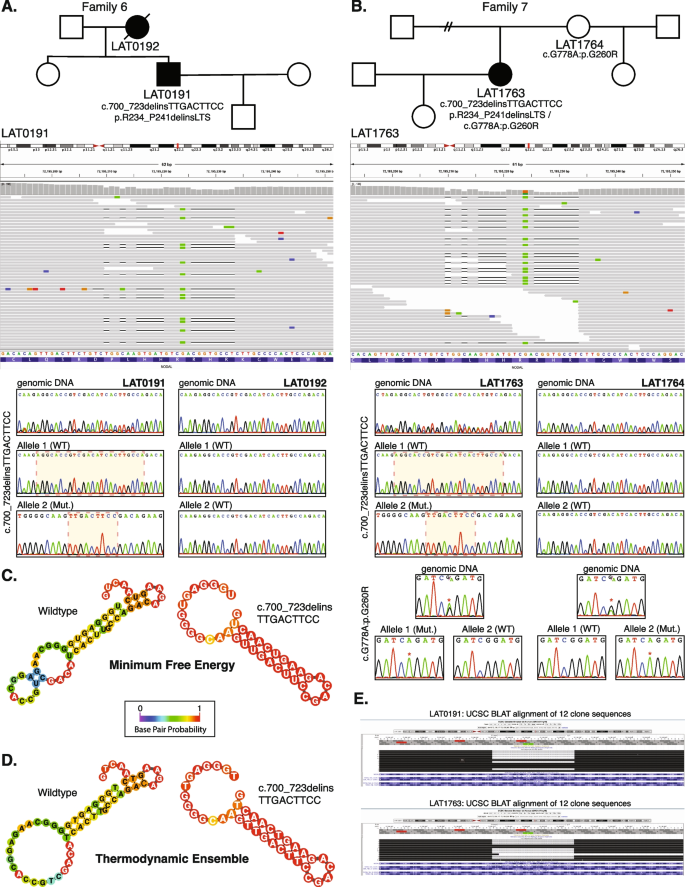
Complex NODAL indel variants in two probands with congenital heart disease. A Pedigree structure for Family 6 (top) with integrative genomics viewer (IGV) view of the heterozygous complex indel variant in NODAL (below). Under the IGV view are Sanger dideoxy sequence traces for the proband, LAT0191 (left) and mother, LAT0192, (right). The Sanger sequence trace panels at top represent PCR amplification products of the variant region from genomic DNA. Individual alleles are not discernable for the proband genomic DNA trace at left. Beneath the genomic DNA results are Sanger sequence traces from cloning the amplified variant region using TA cloning kits as described in “ Methods ”. Top and bottom panels are representative of different populations of clones for each allele. At left, the complex c.700_723delinsTTGACTTCC, p.R234_P241delinsLTS variant is apparent in the Allele 2 (Mut) Sanger trace for the proband, LAT0191. B Pedigree, IGV view, and Sanger traces for Family 7. In addition to the c.700_723delins, p.R234_P241delinsLTS variant, Sanger traces for the nearby c.778G > A:p.G260R variant are shown. Allele 1 in LAT1763 is shown to be WT for the c.700_723 locus, while containing the c.778G > A variant. Allele 2 was shown to contain the c.700_723delins, p.R234_P241delinsLTS variant while being WT at the c.778G > A locus, confirming these variant alleles are in a trans configuration. C , D Secondary structure predictions for WT and mutant RNA using the RNAfold Server ( http://rna.tbi.univie.ac.at/ ) using the minimum free energy ( C ) and thermodynamic ensemble ( D ) functions. Base pair probability is shown using color coding; cooler colors (blue) represent lower probability and warmer colors (red) represent higher probability. Variant RNA has more stable secondary structure as shown at right in C and D . E UCSC genome browser view of the 12 clones sequenced for LAT0191 and LAT1763, showing populations with the deletion, and with WT sequence. All variant data shown is for NODAL transcript NM_018055.5
The G260R variant observed primarily in Hispanic ancestry heterotaxy subjects
The most common variant in the study was the c.778G > A, G260R variant, detected in 17/31 CHD probands, of which all but 1 were of known Hispanic ancestry (Additional file 2 : Table S7). To compare frequencies of this G260R variant allele to that observed in the “normotypical” population ( https://gnomad.broadinstitute.org/variant/10-72195155-C-T?dataset=gnomad_r2_1 ), we focused on the largest cohort of 321 unrelated probands with laterality CHD that were recruited consecutively (Additional file 2 : Table S8). Of these, 111 self-identified as fully or partially Hispanic (35%). The NODAL c.778G > A, G260R variant is by far most common in Hispanic patients, with an allele frequency of 0.00219 (76/34,592 exomes in gnomAD), with no homozygotes observed. Given this frequency, we would expect ≤ 1 G260R variant in the subcohort of 111 Hispanic laterality probands. However, we identified 10 Hispanic cases within this subcohort with a NODAL G260R variant allele (10/111 = 0.090), and two of these cases were biallelic for the G260R variant. That gives an odds ratio of 26.0 (95%CI 13.9–48.4, p < 0.0001) for the cohort. Moreover, if limiting the Hispanic subcohort to the most commonly reported NODAL -associated laterality defects (DTGA, CCTGA, DILV, other L-ventricular looping, and heterotaxy right atrial isomerism type in the cohort), 10 had the G260R variant (10/64 = 15.6%, 12/128 alleles). So, in that group of defects, the odds ratio comparing to the gnomAD population for the G260R variant is 51.4 (95%CI 27.8–95.2, p < 0.0001).
Penetrance and phenotypic variability observed with the heterozygous and homozygous G260R allele
To better understand the phenotypic spectrum associated with potentially pathogenic NODAL alleles, we performed quantitative phenotypic analysis of all NODAL cases with CHD using HPO terms. However atrial septal defect, ventricular septal defect, single ventricle, and secundum atrial septal defect were excluded from the primary analysis due to the nonspecificity of these terms to rare CHD. A supplementary analysis with these terms included is provided for comparison (Additional file 3 : Fig.S1). A gap statistic curve (Additional file 3 : Fig.S2) was used to determine the number of groups to cluster the cases into, and a heatmap of phenotypic similarity scores and clustering was generated using HAC (Fig. 5 ).
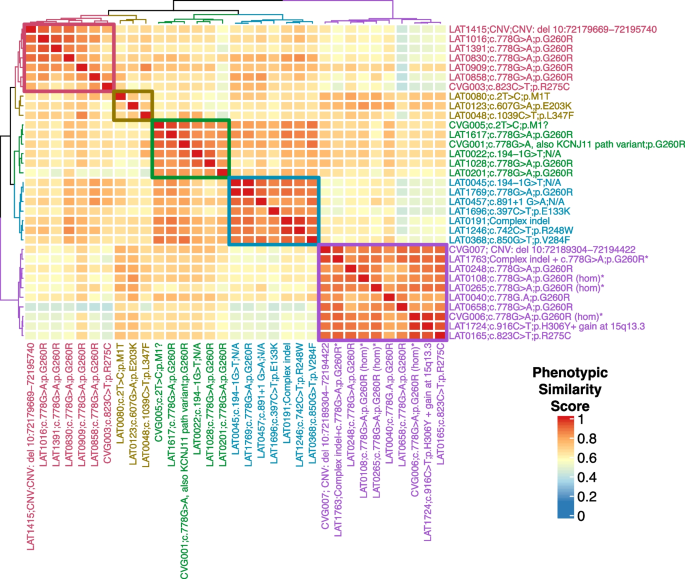
NODAL proband phenotypic similarity heatmap—A heatmap was generated using proband phenotypic similarity scores and ordered based on Hierarchical Agglomerative Clustering of proband phenotypic similarity. Dendrograms showing clusters are present at the left and top sides of the heatmap. Proband IDs and variants are shown at right and bottom, and color coded based on cluster. Five clusters are highlighted by color-coded boxes on the heatmap, from top left diagonally to bottom right: red, olive, green, teal, and purple. The 4 probands with biallelic variation group in one cluster (purple box). A key for phenotypic similarity score based on color (blue corresponding to a lower score and red corresponding to a higher score) is shown at bottom right. An asterisk denotes probands with biallelic variants in NODAL
The phenotypic similarity scores among all clusters were consistently high. Notably, probands with homozygous G260R variants and the compound heterozygous case, LAT1763, formed a distinct cluster (purple group) due to a consistent phenotype: heterotaxy, right atrial isomerism/asplenia type. This group exhibited atrioventricular connection abnormalities, D-MGA, and pulmonary stenosis/atresia. A grid visualization of HPO annotated proband phenotypes (Additional file 3 : Fig.S1A) highlights the severity of the phenotypic spectrum in this cluster. Heterozygous alleles displayed greater phenotypic variability across clusters (Fig. 5 ), with no unaffected cases reported for biallelic predicted deleterious NODAL variants, suggesting potential complete penetrance. Conversely, seven families with heterozygous G260R showed reduced penetrance (Fig. 1 A, Additional file 2 : Table S7).
The red and olive clusters shared higher phenotypic similarity, primarily featuring DTGA (90%) and DORV (70%) without pulmonary artery/valve atresia. The red cluster differentiated by a prevalence of straddling atrioventricular valve and hypoplastic aortic arch with coarctation of the aorta (57.1%). Notably, 66.7% of probands in the olive cluster exhibited a heterotaxy phenotype. The green and teal clusters were closely related phenotypically, with LTGA (92.3%), L-looping of the right ventricle with discordant atrioventricular connection (84.6%), and CCTGA (76.9%) as predominant features. The green cluster further stood out with pulmonary artery/valve atresia (100%) and dextrocardia as a majority feature (66.7%) (Fig. 5 ).
Heterozygous CNV deletions spanning NODAL in laterality defects
Structural genomic variation spanning NODAL was assessed in this laterality defect cohort using XHMM and HMZDelFinder CNV detection tools by comparison of ES read depth data [ 17 , 18 ]. A high-confidence heterozygous deletion was observed in LAT1415 (Fig. 6 A, B), who presented with tricuspid atresia, straddling mitral valve (MV), DORV, D-MGA, and severe coarctation of the aorta. ddPCR confirmed paternal inheritance, with the father having no CHD history (Additional file 3 : Fig.S3).
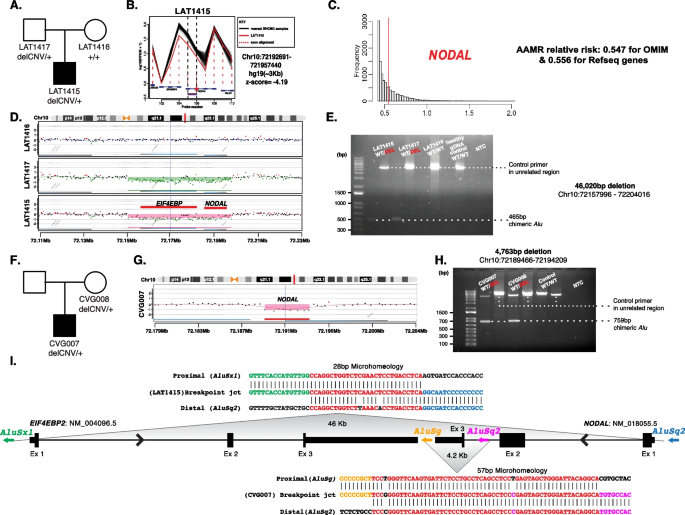
NODAL heterozygous copy number variant (CNV) deletion alleles in laterality defect cases. A Proband LAT1415 pedigree (Family 30) (delCNV/+ heterozygous, +/+ WT homozygous). B HMZDelFinder analyses detected a CNV deletion at the 5′ start of NODAL using ES read count data (RPKM); red vertical dashed lines align to each gene exon, horizontal jagged lines (black: controls; red: deletion CNV subject) show distribution of individual read depth values for that given exon. C The Alu-Alu mediated rearrangement (AAMR) risk score for NODAL and EIF4EBP2 using AluAluCNVpredictor tool. D Family 30 aCGH showed a 46-kb deletion in proband and father. E The 1% agarose gel electrophoresis of PCR products showing the recombinant junction. The junction primer pair was designed to produce an amplicon size of 465 base pairs for deleted alleles resulting from AAMR with the formation of a chimeric Alu . F Family 31 pedigree of proband (CVG007). G Confirmation by aCGH showed a ~4 kb deletion CNV spanning NODAL exon 3. H The gel electrophoresis of PCR products showing the recombinant junction with lighter 700 base pair bands representing heterozygous deleted alleles and more intense bands (~4 kb) representing WT allele. I A schematic representation of NODAL and EIF4EBP2 . Note convergent transcripts for NODAL and EIF4EBP2 (black arrow heads representing gene’s orientation). Breakpoint sequences for NODAL deletions are also shown. The proximal reference sequence and its matching proband breakpoint sequences are shown in green for LAT1415 and orange for CVG007, the distal reference sequence and its matching proband breakpoint sequences are in blue for LAT1415 and purple for CVG007, and microhomology at the junction is shown in red. The 46 kb deletion in LAT1415 presumably results from AAMR between Alu Sx1/ Alu Sq2, whereas the 4 kb deletion in CVG007 proposed to result from AAMR between Alu Sg/ Alu Sq2
The NODAL deletion CNV, spanning the entire gene (46 Kb), was further characterized using high-density aCGH, revealing a high instability score (0.547 for OMIM genes, 0.556 for RefSeq genes) through AluAluCNVpredictor [ 25 ], suggesting susceptibility to Alu-Alu -mediated rearrangement (AAMR; Fig. 6 C). Breakpoint junction analysis (Fig. 6 E) indicated direct-oriented AluSx1/AluSq2 elements, consistent with the predicted AAMR-generated event. This deletion likely resulted from a new mutation in a preceding generation within the family (Fig. 6 I).
Additionally, a maternally inherited single-exon deletion of NODAL was identified in a male patient (CVG0007) with DORV, D-MGA, subaortic stenosis, and severe arch hypoplasia/coarctation (Fig. 6 F–H). His mother had no CHD. This patient was recruited through BG, in which the deletion was initially predicted by CMA testing. Breakpoint junction analysis revealed an AAMR event involving directly oriented AluSg/AluSq2 (Fig. 6 I).
NODAL is a key signaling molecule that plays a pivotal role during embryonic patterning, axis formation, and germ layer specification during early developmental stages. NODAL signaling is also known to be involved in the maintenance of human embryonic stem cell pluripotency and differentiation into specific cell types in a context-dependent manner, and regulation of L-R lineage determination [ 26 , 27 ]. Several animal-model studies including mouse and zebrafish, as well as misexpression studies in chick and Xenopus, have demonstrated the influence of NODAL signaling in heart development along with L-R axis determination [ 28 , 29 ]. To further our understanding of NODAL variants and associated phenotypes, here we investigated NODAL variants in 31 families with CHD (Fig. 1 ). Overall, NODAL variants were observed in 6.5% of the laterality defects cohort (21/321), and in 9% (10/111) of Hispanic patients in the cohort, and in 15.6% (10/64) of Hispanic patients in that cohort with NODAL -associated laterality defects; signifying the contribution of NODAL variation to CHD. Of note, probands in 4 families had biallelic NODAL variant alleles (Fig. 1 B) and one family had one laterality CHD proband (LAT016) with a de novo NODAL SNV and a sibling with CHD associated with a de novo del22q11.2 CNV (Family 18, Fig. 1 A). The latter family speaks to both the rates of structural variant mutagenesis in sporadic birth defects and the need to investigate all CHD probands by genomic studies [ 30 ].
Although there was a spectrum of CHD lesions associated with NODAL variants, three unique patterns were clear. First, all CHD lesions included MGA of some type, whether it be D-transposed or malposed, L-transposed or malposed, or with an anterior aorta and pulmonary atresia in which the exact malposed relationship could not be discerned. This is a critical finding; in that it suggests that CHD lesions with great artery malposition are within the “laterality defect” classification. Historically DTGA, DORV with malposed great arteries, and sometimes CCTGA are often instead classified as conotruncal defects. Despite this, those three lesions are virtually never seen in 22q11.2 deletion syndrome, which is by far the most common cause of conotruncal defects. The lesions tricuspid atresia and DILV have poorly understood genetic etiologies, but multiple cases of both lesions, when present with malposed great arteries, were seen in the cohort. Perhaps separating those with malposed great arteries and those with normal great arteries will help further genetic understanding. Understanding these lesions’ more optimal classification as laterality defects may assist practitioners in pursuing indicated genetic testing, will help variant prioritization and clinical interpretation, and will better guide familial genetic counseling.
Second, almost half of the cohort had left ventricular looping, which is rare even in those with CHD (~2% of CHD [ 31 ]), and 14.1% of the study cohort with left ventricular looping had a NODAL variant. This is logical given NODAL’s critical role in L-R axis patterning. However, in a clinical setting this strong association has not been well appreciated and is even stronger that the most commonly known association between 22q11.2 deletion syndrome and simple tetralogy of Fallot (in which ~8% have 22q11.2 deletion).
Third, NODAL variants when associated with heterotaxy were exclusively associated with either asplenia syndrome/right atrial isomerism ( n = 10) or in two cases with visceral/bronchial/atrial situs inversus with CHD. Right atrial isomerism and left atrial isomerism are often grouped together as similar lesions and even share the same Van Praagh nomenclature of “atrial situs ambiguous”. However, the distinct lack of left atrial isomerism cases in this NODAL cohort and in published literature suggest that genetically and etiologically, these two conditions are distinct and should be evaluated differently when studying genetic mechanisms and inheritance.
The prevalence of NODAL variants we observed is relatively high compared to that reported from the Pediatric Cardiac Genomics Consortium (PCGC) CHD cohort where NODAL variants were identified in only four cases of 2871 CHD probands (Additional file 2 : Tables S9 and S10) [ 32 ]. However, this report was limited to de novo and recessive variant alleles, did not include copy number variants, had a heterogeneous collection of CHD, had a reporting threshold too restrictive to report the G260R variant, and only included 280 patients of Hispanic ancestry (9.8% of the reported). If one limits the study to the 523 laterality CHD cases, NODAL variants account for 0.76% of cases. In our 321-person laterality CHD cohort with 35% Hispanic ethnicity, if the G260R variant and copy number variants are not included, the yield would be much less, with only 10 cases with a NODAL variant (3.1%). Of note, the phenotypes within the PCGC cohort are completely consistent with our observations, with one simple DTGA, one DORV with D-MGA, and two cases of CCTGA (Additional file 2 : Table S10).
The identified NODAL variants in our cohort include eleven SNVs (8 missense, 2 splice site, and 1 nonsense), one recurrent complex indel variant (p.R234_P241delinsLTS), and two CNV deletion alleles (one whole NODAL gene deletion and one exon 3 deletion). Among the identified SNVs, two (p.M1T, p.L347F) were previously unreported variant alleles that have never been associated with laterality defects and are strongly predicted as disease causing using in silico tools.
The complex indel variant c.700_723delins, p.R234_P241delinsLTS, consisting of a 24-base deletion with a 9 base insertion affects the likely cleavage site of the NODAL protein, affecting its recognition by proprotein convertases and impairing protein maturation. While the exact furin cleavage site in NODAL has not been described to our knowledge, the Arginine-Histidine-Arginine-Arginine (RHRR) sequence, specifically between amino acids 234 and 237 in the precursor form of the protein, represents the only amino acid sequence in NODAL that matches the RXXR motif furin cleaves (Additional file 3 : Fig. S4). Secondary structure predictions suggest that the variant may also alter the RNA molecule’s folding and stability, potentially impacting gene expression and developmental processes. These structural changes may affect the accessibility of the RNA molecule to other molecules involved in its processing and function. Overall, the complex allele is likely to have significant effects on RNA and protein structure, as well as gene function, contributing to the observed phenotypic abnormalities associated with laterality defects. However, expression data on cell lines expressing this mutation would be required to confirm changes in gene expression.
In the current cohort, the G260R variant was found in a high prevalence. Four probands with biallelic variation, three homozygotes and one compound heterozygote, were identified. The population specificity of the G260R variant to Hispanics may be due to founder effects resulting from genetic drift. Although specific country of origin ancestry is not available for most patients in this study (only available for 4 of the Hispanic patients which were Mexican), the largest proportion of Hispanics in Texas are of Mexican heritage. None of the probands were known to be related. While formal testing for interrelatedness was not performed, detailed three plus generation family histories were collected both for the study and in the clinical setting and did not suggest any overlap. Additionally, there were no common surnames across the families. Moreover, using an in house tool analyzing the exome data we checked the absence of heterozygosity (AOH)/runs of homozygosity (ROH) regions, especially around this locus, for all the probands with NODAL variants in our study. The AOH/ROH data flanking the G260R locus further supports the low potential of consanguinity in our cohort, given that no AOH regions were observed in this region. However, the detection of this variant in unaffected parents suggests that the variant may not always lead to the development of the condition when in heterozygous state, indicating incomplete penetrance. These findings are consistent with the low recurrence risk and complex inheritance pattern observed in most sporadic cases of laterality defects, which suggests that multiple genetic and environmental factors may contribute to the development of the condition. In other words, the level of NODAL function, due to both genetic and environmental perturbations, may govern penetrance and phenotypic severity of CHD phenotypes. The complete penetrance and consistency of phenotype observed for probands with biallelic predicted deleterious variation in NODAL supports this hypothesis.
One genetic factor that may affect the penetrance of the G260R allele in certain populations is modifying background genetic variation, which is supported by the observation of higher heterotaxy incidence in the African American and Hispanic populations [ 33 ]. The same study also pointed to another factor affecting observed heterotaxy rates, namely diabetes [ 33 ], which may impact penetrance of heterotaxy phenotypes through physiologic factors such as exposure of the developing fetus to insulin [ 34 ]. Of note, penetrance can be influenced by ancestry-specific haplotypes in congenital scoliosis associated with developmental hemivertebrae defects of the spine [ 35 ].
G260R was originally described in 2009 by Mohapatra et al. as pathogenic and causative for heterotaxy with reduced penetrance, variable expressivity, and predominantly affecting Hispanic individuals. Since then, G260R has been found in several individuals with CHD and/or heterotaxy across multiple centers ( https://www.ncbi.nlm.nih.gov/clinvar/variation/8269/ ). In ClinVar, G260R has been classified as “conflicting interpretations of pathogenicity” based on the American College of Medical Genetics and Genomics (ACMG) criteria (ClinVar Accession: VCV000008269.9). It is noteworthy that the G260R variant demonstrates a CADD score of 25.5 and a REVEL score of 0.793, both indicative of its potential pathogenic impact. However, the reclassification of the NODAL G260R allele to an ACMG classification of LP, as proposed in this study, should be considered within the context of our laterality defects cohort. The high prevalence of this variant in our cohort, along with the presence of biallelic cases within our study, supports this reclassification. Furthermore, the implications of this reclassification should be considered in the broader clinical and genetic counseling context, including the potential for preimplantation genetic testing for LP variants.
Quantitative phenotypic analysis showed high phenotypic similarity score between all clusters suggesting no specific genotypic-phenotypic correlations. However, all biallelic cases with G260R variant were shown to have the highest phenotypic similarity between probands suggesting the most consistent phenotype (Fig. 5 and Additional file 2 : Table S6). This finding provides evidence for a genotypic-phenotypic correlation and an allele-specific gene dosage model [ 36 ]. However, it is important to note that the absence of such a correlation among individuals without the variant does not necessarily indicate the absence of other genetic factors influencing the phenotype. It is also important to consider other factors that may contribute to the observed phenotype, such as environmental factors and epigenetic modifications.
We report two unrelated laterality defect cases whose underlying disease-causing mutations were NODAL CNV deletion alleles: a whole gene deletion and a single exon deletion. The CNV deletion alleles were shown to most likely have formed due to AAMR events, which is consistent with the high relative gene/genomic instability score (0.547 for OMIM genes and 0.556 for RefSeq genes) observed for NODAL using the AluAluCNVpredictor [ 25 ]. These observations suggest that NODAL CNV deletions should be considered in genomic diagnostics, genetic counseling, and testing for laterality defects.
Conclusions
Collectively, our findings suggest that assessment of all variant types (SNV, indel, and CNV) in heterotaxy cases can increase molecular diagnosis rates in CHD cases and that allele-specific gene dosage can be an important contributor to penetrance and variable expression of CHD. We confirm that rare variants in NODAL contribute to the development of heterotaxy spectrum congenital heart defects [ 13 ] and provide evidence that the population specificity of these variants should be taken into consideration in genetic counseling and clinical genomic testing for this condition. Moreover, our study uncovers unreported NODAL mutations and mutation types in association with laterality defects, enabling an allelic series that furthers our understanding of the biological perturbations and genetic pathobiology underlying laterality defects. These findings have important implications for the diagnosis and treatment of human laterality defects.
Availability of data and materials
The datasets used and/or analyzed during the current study are available in the supplementary data.
Abbreviations
- Congenital heart disease
Double outlet right ventricle
Atrioventricular canal defect
Transposition of the great arteries
Left-lateral plate mesoderm
Right-lateral plate mesoderm
Pathogenic/likely pathogenic
Clinical microarray
Baylor Genetics
1000 Genomes Project
Atherosclerosis Risk in Communities Study Database
Combined Annotation Dependent Depletion
Luria-Bertani
Hierarchical Agglomerative Clustering
Droplet digital PCR
Array comparative genomic hybridization
Long-range PCR
Congenitally corrected transposition of the great arteries
Levo-transposition of the great arteries
Dextro-transposition of the great arteries
D-malposed great arteries
Pulmonary stenosis
Arteriovenous malformations
Vesicoureteral reflux
Mitral valve
Alu - Alu Mediated rearrangement
Pediatric Cardiac Genomics Consortium
American College of Medical Genetics and Genomics
Moran R, Robin NH. Congenital heart defects. In: Emery and Rimoin’s principles and practice of medical genetics. 2013. p. 1–51.
Andersen TA, Troelsen KDLL, Larsen LA. Of mice and men: molecular genetics of congenital heart disease. Cell Mol Life Sci. 2014;71(8):1327–52. Available from: https://link.springer.com/article/10.1007/s00018-013-1430-1 . Cited 2023 Feb 17.
Article CAS PubMed Google Scholar
Hoffman JIE. Incidence of congenital heart disease: II. Prenatal incidence. Pediatr Cardiol. 1995;16(4):155–65. Available from: https://pubmed.ncbi.nlm.nih.gov/7567659/ . Cited 2023 Feb 16.
Tortigue M, Nield LE, Karakachoff M, McLeod CJ, Belli E, Babu-Narayan SV, et al. Familial recurrence patterns in congenitally corrected transposition of the great arteries: an international study. Circ Genom Precis Med. 2022;15(3):E003464. Available from: https://pubmed.ncbi.nlm.nih.gov/35549293/ . Cited 2023 Apr 30.
Restivo A, Piacentini G, Placidi S, Saffirio C, Marino B. Cardiac outflow tract: a review of some embryogenetic aspects of the conotruncal region of the heart. Anat Rec A Discov Mol Cell Evol Biol. 2006;288(9):936–43. Available from: https://pubmed.ncbi.nlm.nih.gov/16892424/ . Cited 2023 Apr 30.
Article PubMed Google Scholar
D’Alessandro LCA, Latney BC, Paluru PC, Goldmuntz E. The phenotypic spectrum of ZIC3 mutations includes isolated d-transposition of the great arteries and double outlet right ventricle. Am J Med Genet A. 2013;161A(4):792–802. Available from: https://pubmed.ncbi.nlm.nih.gov/23427188/ . Cited 2023 Apr 30.
Brandler WM, Morris AP, Evans DM, Scerri TS, Kemp JP, Timpson NJ, et al. Common variants in left/right asymmetry genes and pathways are associated with relative hand skill. PLoS Genet. 2013;9(9):e1003751. Available from: https://pubmed.ncbi.nlm.nih.gov/24068947/ . Cited 2023 Apr 30.
Article CAS PubMed PubMed Central Google Scholar
Belmont JW, Mohapatra B, Towbin JA, Ware SM. Molecular genetics of heterotaxy syndromes. Curr Opin Cardiol. 2004;19(3):216–20. Available from: https://pubmed.ncbi.nlm.nih.gov/15096953/ . Cited 2023 Feb 17.
Kosaki K, Casey B. Genetics of human left-right axis malformations. Semin Cell Dev Biol. 1998;9(1):89–99. Available from: https://pubmed.ncbi.nlm.nih.gov/9572118/ . Cited 2023 Feb 17.
Wells JR, Padua MB, Ware SM. The genetic landscape of cardiovascular left–right patterning defects. Curr Opin Genet Dev. 2022;75:101937.
Shiratori H, Hamada H. The left-right axis in the mouse: from origin to morphology. Development. 2006;133(11):2095–104. Available from: https://pubmed.ncbi.nlm.nih.gov/16672339/ . Cited 2023 Feb 16.
Shiratori H, Hamada H. TGFβ signaling in establishing left-right asymmetry. Semin Cell Dev Biol. 2014;32:80–4. Available from: https://pubmed.ncbi.nlm.nih.gov/24704359/ . Cited 2023 Feb 16.
Mohapatra B, Casey B, Li H, Ho-Dawson T, Smith L, Fernbach SD, et al. Identification and functional characterization of NODAL rare variants in heterotaxy and isolated cardiovascular malformations. Hum Mol Genet. 2009;18(5):861–71. Available from: https://academic.oup.com/hmg/article/18/5/861/615464 . Cited 2023 Feb 17.
Reid JG, Carroll A, Veeraraghavan N, Dahdouli M, Sundquist A, English A, et al. Launching genomics into the cloud: deployment of Mercury, a next generation sequence analysis pipeline. BMC Bioinformatics. 2014;15(1):1–11. Available from: https://bmcbioinformatics.biomedcentral.com/articles/10.1186/1471-2105-15-30 . Cited 2023 May 11.
Article Google Scholar
Li AH, Hanchard NA, Azamian M, D’Alessandro LCA, Coban-Akdemir Z, Lopez KN, et al. Genetic architecture of laterality defects revealed by whole exome sequencing. Eur J Hum Genet. 2019;27(4):563–73. Available from: https://www.nature.com/articles/s41431-018-0307-z . Cited 2023 Feb 17.
Yang A, Alankarage D, Cuny H, Ip EKK, Almog M, Lu J, et al. CHDgene: a curated database for congenital heart disease genes. Circ Genom Precis Med. 2022;15(3):E003539. Available from: https://www.ahajournals.org/doi/abs/10.1161/CIRCGEN.121.003539 . Cited 2024 Jan 28.
Fromer M, Moran JL, Chambert K, Banks E, Bergen SE, Ruderfer DM, et al. Discovery and statistical genotyping of copy-number variation from whole-exome sequencing depth. Am J Hum Genet. 2012;91(4):597–607. Available from: https://pubmed.ncbi.nlm.nih.gov/23040492/ . Cited 2023 Feb 18.
Gambin T, Akdemir ZC, Yuan B, Gu S, Chiang T, Carvalho CMB, et al. Homozygous and hemizygous CNV detection from exome sequencing data in a Mendelian disease cohort. Nucleic Acids Res. 2017;45(4):1633–48. Available from: https://pubmed.ncbi.nlm.nih.gov/27980096/ . Cited 2023 Feb 18.
CAS PubMed Google Scholar
Zhang C, Jolly A, Shayota BJ, Mazzeu JF, Du H, Dawood M, et al. Novel pathogenic variants and quantitative phenotypic analyses of Robinow syndrome: WNT signaling perturbation and phenotypic variability. HGG Adv. 2021;3(1):100074. Available from: https://pubmed.ncbi.nlm.nih.gov/35047859/ . Cited 2023 Apr 14.
PubMed PubMed Central Google Scholar
Greene D, Richardson S, Turro E. ontologyX: a suite of R packages for working with ontological data. Bioinformatics. 2017;33(7):1104–6. Available from: https://pubmed.ncbi.nlm.nih.gov/28062448/ . Cited 2023 Apr 14.
Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32(18):2847–9. Available from: https://pubmed.ncbi.nlm.nih.gov/27207943/ . Cited 2023 Apr 14.
Liu Q, Grochowski CM, Bi W, Lupski JR, Stankiewicz P. Quantitative assessment of parental somatic mosaicism for CNV deletions. Curr Protoc Hum Genet. 2020;106(1):e99. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7138410/ . Cited 2023 Jun 6.
Carvalho CMB, Zhang F, Liu P, Patel A, Sahoo T, Bacino CA, et al. Complex rearrangements in patients with duplications of MECP2 can occur by fork stalling and template switching. Hum Mol Genet. 2009;18(12):2188. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2685756/ . Cited 2023 Jun 6.
Van Praagh R. Terminology of congenital heart disease. Glossary and commentary. Circulation. 1977;56(2):139–43. https://doi.org/10.1161/01.cir.56.2.139 .
Song X, Beck CR, Du R, Campbell IM, Coban-Akdemir Z, Gu S, et al. Predicting human genes susceptible to genomic instability associated with Alu/Alu-mediated rearrangements. Genome Res. 2018;28(8):1228–42. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6071635/ . Cited 2023 May 21.
Yook JY, Kim MJ, Son MJ, Lee S, Nam Y, Han YM, et al. Combinatorial activin receptor-like kinase/Smad and basic fibroblast growth factor signals stimulate the differentiation of human embryonic stem cells into the cardiac lineage. https://home.liebertpub.com/scd . 2011;20(9):1479–90. Available from: https://www.liebertpub.com/doi/10.1089/scd.2010.0392 . Cited 2023 Mar 5.
Smith JR, Vallier L, Lupo G, Alexander M, Harris WA, Pedersen RA. Inhibition of Activin/Nodal signaling promotes specification of human embryonic stem cells into neuroectoderm. Dev Biol. 2008;313(1):107–17.
Brennan J, Norris DP, Robertson EJ. Nodal activity in the node governs left-right asymmetry. Genes Dev. 2002;16(18):2339–44. Available from: http://genesdev.cshlp.org/content/16/18/2339.full . Cited 2023 Mar 5.
Lowe LA, Yamada S, Kuehn MR. Genetic dissection of nodal function in patterning the mouse embryo. Development. 2001;128(10):1831–43. Available from: https://journals.biologists.com/dev/article/128/10/1831/41309/Genetic-dissection-of-nodal-function-in-patterning . Cited 2023 Mar 5.
Bi W, Probst FJ, Wiszniewska J, Plunkett K, Roney EK, Carter BS, et al. Co-occurrence of recurrent duplications of the DiGeorge syndrome region on both chromosome 22 homologues due to inherited and de novo events. J Med Genet. 2012;49(11):681–8. Available from: https://pubmed.ncbi.nlm.nih.gov/23042811/ . Cited 2023 May 21.
Kowalik E. Management of congenitally corrected transposition from fetal diagnosis to adulthood. Expert Rev Cardiovasc Ther. 2023. Available from: https://pubmed.ncbi.nlm.nih.gov/37143366/ . Cited 2023 Jun 11.
Jin SC, Homsy J, Zaidi S, Lu Q, Morton S, Depalma SR, et al. Contribution of rare inherited and de novo variants in 2,871 congenital heart disease probands. Nat Genet. 2017;49(11):1593–601. Available from: https://pubmed.ncbi.nlm.nih.gov/28991257/ . Cited 2023 Apr 14.
Lopez KN, Marengo LK, Canfield MA, Belmont JW, Dickerson HA. Racial disparities in heterotaxy syndrome. Birth Defects Res A Clin Mol Teratol. 2015;103(11):941–50. Available from: https://pubmed.ncbi.nlm.nih.gov/26333177/ . Cited 2023 Apr 14.
Luo Z, Xu L, Lu J, Shen Y, Tang Y, Wang X, et al. Down-regulation of the insulin signaling pathway by SHC may correlate with congenital heart disease in Chinese populations. Clin Sci (Lond). 2020;134(3):349–68. Available from: https://pubmed.ncbi.nlm.nih.gov/31971563/ . Cited 2023 Apr 14.
Wu N, Ming X, Xiao J, Wu Z, Chen X, Shinawi M, et al. TBX6 null variants and a common hypomorphic allele in congenital scoliosis. N Engl J Med. 2015;372(4):341–50. Available from: https://pubmed.ncbi.nlm.nih.gov/25564734/ . Cited 2023 May 21.
Duan R, Hijazi H, Gulec EY, Eker HK, Costa SR, Sahin Y, et al. Developmental genomics of limb malformations: allelic series in association with gene dosage effects contribute to the clinical variability. HGG Adv. 2022;3(4):100132. Available from: https://pubmed.ncbi.nlm.nih.gov/36035248/ . Cited 2023 Apr 15.
CAS PubMed PubMed Central Google Scholar
Download references
Acknowledgements
We would like to thank all the families and patients for their participation as well as referring genetic counselors and physicians.
This work was supported in part by the US National Human Genome Research Institute/National Heart Blood Lung Institute jointly funded Baylor Hopkins Center for Mendelian Genomics (UM1HG006542), by National Institutes of Health (NIH) grants to S.A.M. (5RO1 HD039056, 5RO1 HL091771), by the Genomic Research Elucidates the Genetics of Rare disease (GREGoR) program (UM1 HG011758) to J.E.P., J.R.L., and R.A.G., and by the National Institute of Neurological Disorders and Stroke (NINDS R35 NS105078) to J.R.L.. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Author information
Zain Dardas, Jawid M. Fatih, and Angad Jolly contributed equally to this work.
Authors and Affiliations
Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, TX, 77030, USA
Zain Dardas, Jawid M. Fatih, Angad Jolly, Moez Dawood, Haowei Du, Christopher M. Grochowski, Shalini N. Jhangiani, Pengfei Liu, Weimin Bi, Jennifer E. Posey, Donna M. Muzny, Richard A. Gibbs, James R. Lupski & Zeynep Coban-Akdemir
Human Genome Sequencing Center, Baylor College of Medicine, Houston, TX, 77030, USA
Moez Dawood, Shalini N. Jhangiani, Eric Boerwinkle, Donna M. Muzny, Richard A. Gibbs & James R. Lupski
Medical Scientist Training Program, Baylor College of Medicine, Houston, TX, 77030, USA
Moez Dawood
Division of Cardiology, Department of Pediatrics, Texas Children’s Hospital and Baylor College of Medicine, Houston, TX, 77030, USA
Edward G. Jones, Xander H. T. Wehrens & Shaine A. Morris
Cardiovascular Research Institute, Baylor College of Medicine, Houston, TX, 77030, USA
Xander H. T. Wehrens
Department of Integrative Physiology, Baylor College of Medicine, Houston, TX, 77030, USA
Department of Medicine, Baylor College of Medicine, Houston, TX, 77030, USA
Baylor Genetics, Houston, TX, 77021, USA
Pengfei Liu & Weimin Bi
Human Genetics Center, Department of Epidemiology, Human Genetics, and Environmental Sciences, School of Public Health, The University of Texas Health Science Center at Houston, Houston, TX, 77030, USA
Eric Boerwinkle & Zeynep Coban-Akdemir
Texas Children’s Hospital, Houston, Houston, TX, 77030, USA
James R. Lupski
Department of Pediatrics, Baylor College of Medicine, Houston, TX, 77030, USA
You can also search for this author in PubMed Google Scholar
Contributions
Conceptualization: ZD, JRL, ZCA, SAM, Data curation: SNJ, HD, WB, DMM, PL, EB, JEP, RAG, Formal analysis: MD, CMG, ZCA, Funding acquisition: Visualization: JMF, AJ, ZD, Clinical data: SAM, EGJ, XHTW, HPO Analysis: AJ, Writing original draft: Z.D, Writing review and editing: ZD, AJ, JRL, ZCA, SAM. All authors read and approved the final manuscript.
Corresponding authors
Correspondence to Zeynep Coban-Akdemir or Shaine A. Morris .
Ethics declarations
Ethics approval and consent to participate.
The Institutional Review Board of Baylor College of Medicine approved all research study protocols. Written informed consent was obtained from all participating individuals including probands and any available family members from Group 1. For the clinically tested patients (Group 2), a waiver of consent was granted as part of 2 retrospective cohort studies of clinical genetic testing in CHD (IRB approval numbers: H-48014 and H-41191). These approvals were obtained in accordance with the Helsinki Declaration.
Consent for publication
Not applicable.
Competing interests
J.R.L. has stock ownership in 23andMe, is a paid consultant for Genome International, and is a co-inventor on multiple U.S. and European patents related to molecular diagnostics for inherited neuropathies, genomic disorders, eye diseases, and bacterial genomic fingerprinting. The Department of Molecular and Human Genetics at Baylor College of Medicine derives revenue from the chromosomal microarray analysis and clinical genomic sequencing (both ES and WGS) offered in the Baylor Genetics Laboratory ( http://bmgl.com ). J.R.L. serves on the Scientific Advisory Board of BG. The remaining authors declare that they have no competing interests.
Additional information
Publisher’s note.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: table s1..
Molecular, Cohort, and Phenotypic information on all cases with NODAL variants. Table S2. Inclusion criteria and description of included patient groups. Table S3. Detailed clinical information for all 33 CHD cases in the study. Table S4. CHD gene list analyzed in our cohort. Table S5. HPO terms used for all CHD cases in the study.
Additional file 2: Table S6.
DdPCR primer design for the NODAL gene and RPP30 control gene. Table S7. Phenotype for of all cases with p.G260R variation including heterozygous and homozygous cases. Table S8. Allele Frequency comparison of patients with G260R NODAL variant. Table S9. NODAL variation in PCGC cohort. Table S10. Summarized Clinical Information of cases in PCGC Cohort.
Additional file 3: Figure S1.
NODAL Phenotype Grid Comparison – (A) A grid of proband phenotypes was generated using HPO annotated term sets for each proband and ordered based off Hierarchical Agglomerative Clustering of proband phenotypic similarity scores. Probands and variants are labeled at left and color coded by clusters. Colors for each cluster match those displayed in the heatmap. HPO terms are displayed at the bottom of the grid. Within the grid, red denotes presence of a phenotype, while grey denotes absence or lack of clinical data of a phenotype. Frequency for each HPO phenotype in the NODAL cohort is shown by the distribution bar graph at top. An asterisk at the right end of individual proband sample number identifier denotes probands found to have biallelic variants in NODAL . (B) A grid of proband phenotypes was generated using HPO annotated term sets for each proband with atrial septal defect, ventricular septal defect, single ventricle, and secundum atrial septal defect included (bottom) for comparison to the grid of proband phenotypes presented in (A). Colors from the analysis with atrial septal defect, ventricular septal defect, single ventricle, and secundum atrial septal defect removed are preserved to show differences in clustering between the analyses with and without these terms. Figure S2. Gap Statistic Curve – Gap statistic results for hierarchical clustering of the distance matrix generated from the similarity matrix of pairwise proband phenotype similarity scores is shown. The gap statistic is shown on the y-axis and the number of clusters considered is shown on the x-axis. The slope of the curve is steepest before 5 clusters, and so 5 was chosen for the number of clusters to group NODAL proband phenotypes into. Figure S3. Copy number analysis of NODAL by Droplet-digital PCR (ddPCR) for families 30 and 31. (A) The deletion of NODAL was found in the proband (LAT1415) and father (LAT1417) in this pedigree (family 30). Analysis of the mother (LAT1416) and a healthy unrelated control shows normal copy number. (B) The deletion of NODAL exon 3 was found in the proband (CVG007) and mother (CVG008) in this pedigree (family 31). Father’s DNA was not available for testing. Figure S4. Diagrams of the NODAL preproprotein predicted structure highlighting the Furin/PACE4 cleavage site motif. (A) Wild type NODAL tertiary structure model generated by https://alphafold.ebi.ac.uk/entry/Q96S42 , with a zoomed in view at the Furin/PACE4 cleavage site motif (RHRR) in the context of the protein structure. (B) Simplified illustration of the NODAL primary structure of the wild type (top diagram) showing the presence of the Furin/PACE4 cleavage site motif. The deleted amino acids p.R234_P241del (middle diagram), and the mutated p.R234_P241delinsLTS form (bottom diagram) showing the disrupted Furin/PACE4 cleavage site.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/ . The Creative Commons Public Domain Dedication waiver ( http://creativecommons.org/publicdomain/zero/1.0/ ) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
Reprints and permissions
About this article
Cite this article.
Dardas, Z., Fatih, J.M., Jolly, A. et al. NODAL variants are associated with a continuum of laterality defects from simple D-transposition of the great arteries to heterotaxy. Genome Med 16 , 53 (2024). https://doi.org/10.1186/s13073-024-01312-9
Download citation
Received : 26 June 2023
Accepted : 12 March 2024
Published : 03 April 2024
DOI : https://doi.org/10.1186/s13073-024-01312-9
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Laterality defects
- Transposition
- Single ventricle
- Genetic diagnosis
- Structural variation
Genome Medicine
ISSN: 1756-994X
- Submission enquiries: [email protected]
- General enquiries: [email protected]

The use and impact of surveillance-based technology initiatives in inpatient and acute mental health settings: A systematic review
- Find this author on Google Scholar
- Find this author on PubMed
- Search for this author on this site
- ORCID record for Jessica L. Griffiths
- For correspondence: [email protected]
- ORCID record for Katherine R. K. Saunders
- ORCID record for Una Foye
- ORCID record for Anna Greenburgh
- ORCID record for Antonio Rojas-Garcia
- ORCID record for Brynmor Lloyd-Evans
- ORCID record for Sonia Johnson
- ORCID record for Alan Simpson
- Info/History
- Supplementary material
- Preview PDF
Background: The use of surveillance technologies is becoming increasingly common in inpatient mental health settings, commonly justified as efforts to improve safety and cost-effectiveness. However, the use of these technologies has been questioned in light of limited research conducted and the sensitivities, ethical concerns and potential harms of surveillance. This systematic review aims to: 1) map how surveillance technologies have been employed in inpatient mental health settings, 2) identify any best practice guidance, 3) explore how they are experienced by patients, staff and carers, and 4) examine evidence regarding their impact. Methods: We searched five academic databases (Embase, MEDLINE, PsycInfo, PubMed and Scopus), one grey literature database (HMIC) and two pre-print servers (medRxiv and PsyArXiv) to identify relevant papers published up to 18/09/2023. We also conducted backwards and forwards citation tracking and contacted experts to identify relevant literature. Quality was assessed using the Mixed Methods Appraisal Tool. Data were synthesised using a narrative approach. Results: A total of 27 studies were identified as meeting the inclusion criteria. Included studies reported on CCTV/video monitoring (n = 13), Vision-Based Patient Monitoring and Management (VBPMM) (n = 6), Body Worn Cameras (BWCs) (n = 4), GPS electronic monitoring (n = 2) and wearable sensors (n = 2). Twelve papers (44.4%) were rated as low quality, five (18.5%) medium quality, and ten (37.0%) high quality. Five studies (18.5%) declared a conflict of interest. We identified minimal best practice guidance. Qualitative findings indicate that patient, staff and carer perceptions and experiences of surveillance technologies are mixed and complex. Quantitative findings regarding the impact of surveillance on outcomes such as self-harm, violence, aggression, care quality and cost-effectiveness were inconsistent or weak. Discussion: There is currently insufficient evidence to suggest that surveillance technologies in inpatient mental health settings are achieving the outcomes they are employed to achieve, such as improving safety and reducing costs. The studies were generally of low methodological quality, lacked lived experience involvement, and a substantial proportion (18.5%) declared conflicts of interest. Further independent coproduced research is needed to more comprehensively evaluate the impact of surveillance technologies in inpatient settings, including harms and benefits. If surveillance technologies are to be implemented, it will be important to engage all key stakeholders in the development of policies, procedures and best practice guidance to regulate their use, with a particular emphasis on prioritising the perspectives of patients.
Competing Interest Statement
AS and UF have undertaken and published research on BWCs. We have received no financial support from BWC or any other surveillance technology companies. All other authors declare no competing interests.
Clinical Protocols
https://www.crd.york.ac.uk/prospero/display_record.php?RecordID=463993
Funding Statement
This study is funded by the National Institute for Health and Care Research (NIHR) Policy Research Programme (grant no. PR-PRU-0916-22003). The views expressed are those of the author(s) and not necessarily those of the NIHR or the Department of Health and Social Care. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. ARG was supported by the Ramon y Cajal programme (RYC2022-038556-I), funded by the Spanish Ministry of Science, Innovation and Universities.
Author Declarations
I confirm all relevant ethical guidelines have been followed, and any necessary IRB and/or ethics committee approvals have been obtained.
I confirm that all necessary patient/participant consent has been obtained and the appropriate institutional forms have been archived, and that any patient/participant/sample identifiers included were not known to anyone (e.g., hospital staff, patients or participants themselves) outside the research group so cannot be used to identify individuals.
I understand that all clinical trials and any other prospective interventional studies must be registered with an ICMJE-approved registry, such as ClinicalTrials.gov. I confirm that any such study reported in the manuscript has been registered and the trial registration ID is provided (note: if posting a prospective study registered retrospectively, please provide a statement in the trial ID field explaining why the study was not registered in advance).
I have followed all appropriate research reporting guidelines, such as any relevant EQUATOR Network research reporting checklist(s) and other pertinent material, if applicable.
Data Availability
The template data extraction form is available in Supplementary 1. MMAT quality appraisal ratings for each included study are available in Supplementary 2. All data used is publicly available in the published papers included in this review.
View the discussion thread.
Supplementary Material
Thank you for your interest in spreading the word about medRxiv.
NOTE: Your email address is requested solely to identify you as the sender of this article.

Citation Manager Formats
- EndNote (tagged)
- EndNote 8 (xml)
- RefWorks Tagged
- Ref Manager
- Tweet Widget
- Facebook Like
- Google Plus One
- Addiction Medicine (316)
- Allergy and Immunology (617)
- Anesthesia (159)
- Cardiovascular Medicine (2276)
- Dentistry and Oral Medicine (279)
- Dermatology (201)
- Emergency Medicine (370)
- Endocrinology (including Diabetes Mellitus and Metabolic Disease) (798)
- Epidemiology (11573)
- Forensic Medicine (10)
- Gastroenterology (678)
- Genetic and Genomic Medicine (3575)
- Geriatric Medicine (336)
- Health Economics (616)
- Health Informatics (2304)
- Health Policy (913)
- Health Systems and Quality Improvement (863)
- Hematology (335)
- HIV/AIDS (752)
- Infectious Diseases (except HIV/AIDS) (13149)
- Intensive Care and Critical Care Medicine (755)
- Medical Education (359)
- Medical Ethics (100)
- Nephrology (388)
- Neurology (3346)
- Nursing (191)
- Nutrition (506)
- Obstetrics and Gynecology (651)
- Occupational and Environmental Health (645)
- Oncology (1756)
- Ophthalmology (524)
- Orthopedics (209)
- Otolaryngology (284)
- Pain Medicine (223)
- Palliative Medicine (66)
- Pathology (437)
- Pediatrics (1001)
- Pharmacology and Therapeutics (422)
- Primary Care Research (406)
- Psychiatry and Clinical Psychology (3058)
- Public and Global Health (5983)
- Radiology and Imaging (1221)
- Rehabilitation Medicine and Physical Therapy (714)
- Respiratory Medicine (811)
- Rheumatology (367)
- Sexual and Reproductive Health (350)
- Sports Medicine (316)
- Surgery (386)
- Toxicology (50)
- Transplantation (170)
- Urology (142)

IMAGES
VIDEO
COMMENTS
Abstract. The purpose of this study is to critically analyse and evaluate two selected journal articles. The two articles are research papers in the education field; more specifically both ...
The research process for conducting a critical analysis literature review has three phases ; (a) the deconstruction phase in which the individually reviewed studies are broken down into separate discreet data points or variables (e.g., breastfeeding duration, study design, sampling methods); (b) the analysis phase that includes both cross-case ...
Critical Analysis Format is as follows: I. Introduction. Provide a brief overview of the text, object, or event being analyzed. Explain the purpose of the analysis and its significance. Provide background information on the context and relevant historical or cultural factors. II.
Critical appraisal 'The notion of systematic review - looking at the totality of evidence - is quietly one of the most important innovations in medicine over the past 30 years' (Goldacre, Citation 2011, p. xi).These sentiments apply equally to sport and exercise psychology; systematic review or evidence synthesis provides transparent and methodical procedures that assist reviewers in ...
The components of the critical appraisal are the appropriateness of the study design for the research question and a thorough evaluation of important methodological characteristics of this study ...
A critical review (sometimes called a critique, critical commentary, critical appraisal, critical analysis) is a detailed commentary on and critical evaluation of a text. You might carry out a critical review as a stand-alone exercise, or as part of your research and preparation for writing a literature review. The
If the number of cases is too low, a real difference, e.g. between the effects of two medications or in the risk of disease in the presence vs. absence of a given environmental factor, may not be detected. ... Critical analysis of the study's ... How to write the methods section of a research paper. Respir Care. 2004; 49:1229-1232. [Google ...
A research student's first exposure to the literature (a collection of academic research, usually in the form of articles and books) usually involves finding and reading multiple papers, to identify an idea to serve as the basis of his/her thesis. (Please refer to Table 1 for the definitions of this and other key concepts in this paper). Once a ...
This paper describes a broad framework of critical appraisal of published research literature that covers both quantitative and qualitative methodologies. The aim is the heart of a research study. It should be robust, concisely stated and specify a study factor, outcome factor(s) and reference population. Quantitative study designs, including ...
Analysis refers to the assessment, or critique, of the literature (p. 274) The critical reflection Callahan (2014) described is simi-lar to the reflexivity required when conducting a qualitative study (Dodgson, 2019). It is not traditional for authors of literature review research to critically reflect on their assump-tions, beliefs or values ...
Writing Critical Analysis Papers1 A critical analysis paper asks the writer to make an argument about a particular book, essay, movie, etc. The goal is two fold: one, identify and explain the argument that the author is making, and two, provide your own argument about that argument. One of the key directions of these assignments is often
A critical analysis is an argument about a particular piece of media. There are typically two parts: (1) identify and explain the argument the author is making, and (2), provide your own argument about that argument. Your instructor may have very specific requirements on how you are to write your critical analysis, so make sure you read your ...
Does the title precisely state the subject of the paper? Abstract. Read the statement of purpose in the abstract. Does it match the one in the introduction? Acknowledgments. Could the source of the research funding have influenced the research topic or conclusions? Introduction. Check the sequence of statements in the introduction.
The primary focus of this study is to critically analyse two academic papers published in the educational field in terms of the validity and reliability of their methods of data collection and analysis, research design, and ethical implications. This is done in an attempt to demonstrate the valid procedure of conducting a research paper as a general aim for the current study.
Critical Analysis of the Content. ... Primary sources are the raw material of the research process. Secondary sources are based on primary sources. For example, if you were researching Konrad Adenauer's role in rebuilding West Germany after World War II, Adenauer's own writings would be one of many primary sources available on this topic. ...
This paper is to pinpoint their weaknesses and strengths. The two chosen papers are also critiqued from various perspectives such as methods of data collection and analysis, research layout and organization, validity and reliability of the data collected, sampling issue and ethical considerations.
critiquing the literature, critical analysis, reviewing the literature, evaluation and appraisal of the literature which are in essence the same thing (Bassett and Bassett, 2003). Terminology in research can be confusing for the novice research reader where a term like 'random' refers to an organized manner of selecting items or participants ...
Below are nine organizational and writing tips to help you craft the best possible critical analysis essay. 1. Read Thoroughly and Carefully. You will need to accurately represent an author's point of view and techniques. Be sure you truly understand them before you begin the writing process. 2.
1 Important points to consider when critically evaluating published research papers. Simple review articles (also referred to as 'narrative' or 'selective' reviews), systematic reviews and meta-analyses provide rapid overviews and 'snapshots' of progress made within a field, summarising a given topic or research area.
A critical analysis of your sources is key to creating a quality literature review, and keeping your research question in mind as you read the literature will ensure that you are on track. ... A literature review has a format similar to other scholarly papers. It contains an introduction, body and conclusion, but is focused exclusively on the ...
The final section of a journal analysis paper should bring your thoughts together into a coherent assessment of the value of the research study. This section is where the narrative flow transitions from analyzing specific elements of the article to critically evaluating the overall study.
After deleting duplicates from two databases, the research team performed a comprehensive analysis of the 75 papers that resulted from this approach. An open coding system was used in this study to make it easier to examine the collected documents. ... Development of critical reading skills in research in two differentiated groups: Aguilar, HC ...
Habitat change and fragmentation are the primary causes of biodiversity loss worldwide. Recent decades have seen a surge of funding, published papers and citations in the field as these threats to biodiversity continue to rise. However, how research directions and agenda are evolving in this field remains poorly understood. In this study, we examined the current state of research on habitat ...
A critical analysis of the included studies revealed issues related to research design, varying from data collected from heterogeneous healthcare settings and diverse types of MMs to the type of analyses completed (e.g., qualitative, mixed methods), to the strength of conclusions drawn from a few studies' results (e.g., correlational, or causal).
NODAL signaling plays a critical role in embryonic patterning and heart development in vertebrates. Genetic variants resulting in perturbations of the TGF-β/NODAL signaling pathway have reproducibly been shown to cause laterality defects in humans. To further explore this association and improve genetic diagnosis, the study aims to identify and characterize a broader range of NODAL variants ...
Background: The use of surveillance technologies is becoming increasingly common in inpatient mental health settings, commonly justified as efforts to improve safety and cost-effectiveness. However, the use of these technologies has been questioned in light of limited research conducted and the sensitivities, ethical concerns and potential harms of surveillance. This systematic review aims to ...