Organizing Your Social Sciences Research Assignments
- Annotated Bibliography
- Analyzing a Scholarly Journal Article
- Group Presentations
- Dealing with Nervousness
- Using Visual Aids
- Grading Someone Else's Paper
- Types of Structured Group Activities
- Group Project Survival Skills
- Leading a Class Discussion
- Multiple Book Review Essay
- Reviewing Collected Works
- Writing a Case Analysis Paper
- Writing a Case Study
- About Informed Consent
- Writing Field Notes
- Writing a Policy Memo
- Writing a Reflective Paper
- Writing a Research Proposal
- Generative AI and Writing
- Acknowledgments

Definition and Introduction
Journal article analysis assignments require you to summarize and critically assess the quality of an empirical research study published in a scholarly [a.k.a., academic, peer-reviewed] journal. The article may be assigned by the professor, chosen from course readings listed in the syllabus, or you must locate an article on your own, usually with the requirement that you search using a reputable library database, such as, JSTOR or ProQuest . The article chosen is expected to relate to the overall discipline of the course, specific course content, or key concepts discussed in class. In some cases, the purpose of the assignment is to analyze an article that is part of the literature review for a future research project.
Analysis of an article can be assigned to students individually or as part of a small group project. The final product is usually in the form of a short paper [typically 1- 6 double-spaced pages] that addresses key questions the professor uses to guide your analysis or that assesses specific parts of a scholarly research study [e.g., the research problem, methodology, discussion, conclusions or findings]. The analysis paper may be shared on a digital course management platform and/or presented to the class for the purpose of promoting a wider discussion about the topic of the study. Although assigned in any level of undergraduate and graduate coursework in the social and behavioral sciences, professors frequently include this assignment in upper division courses to help students learn how to effectively identify, read, and analyze empirical research within their major.
Franco, Josue. “Introducing the Analysis of Journal Articles.” Prepared for presentation at the American Political Science Association’s 2020 Teaching and Learning Conference, February 7-9, 2020, Albuquerque, New Mexico; Sego, Sandra A. and Anne E. Stuart. "Learning to Read Empirical Articles in General Psychology." Teaching of Psychology 43 (2016): 38-42; Kershaw, Trina C., Jordan P. Lippman, and Jennifer Fugate. "Practice Makes Proficient: Teaching Undergraduate Students to Understand Published Research." Instructional Science 46 (2018): 921-946; Woodward-Kron, Robyn. "Critical Analysis and the Journal Article Review Assignment." Prospect 18 (August 2003): 20-36; MacMillan, Margy and Allison MacKenzie. "Strategies for Integrating Information Literacy and Academic Literacy: Helping Undergraduate Students make the most of Scholarly Articles." Library Management 33 (2012): 525-535.
Benefits of Journal Article Analysis Assignments
Analyzing and synthesizing a scholarly journal article is intended to help students obtain the reading and critical thinking skills needed to develop and write their own research papers. This assignment also supports workplace skills where you could be asked to summarize a report or other type of document and report it, for example, during a staff meeting or for a presentation.
There are two broadly defined ways that analyzing a scholarly journal article supports student learning:
Improve Reading Skills
Conducting research requires an ability to review, evaluate, and synthesize prior research studies. Reading prior research requires an understanding of the academic writing style , the type of epistemological beliefs or practices underpinning the research design, and the specific vocabulary and technical terminology [i.e., jargon] used within a discipline. Reading scholarly articles is important because academic writing is unfamiliar to most students; they have had limited exposure to using peer-reviewed journal articles prior to entering college or students have yet to gain exposure to the specific academic writing style of their disciplinary major. Learning how to read scholarly articles also requires careful and deliberate concentration on how authors use specific language and phrasing to convey their research, the problem it addresses, its relationship to prior research, its significance, its limitations, and how authors connect methods of data gathering to the results so as to develop recommended solutions derived from the overall research process.
Improve Comprehension Skills
In addition to knowing how to read scholarly journals articles, students must learn how to effectively interpret what the scholar(s) are trying to convey. Academic writing can be dense, multi-layered, and non-linear in how information is presented. In addition, scholarly articles contain footnotes or endnotes, references to sources, multiple appendices, and, in some cases, non-textual elements [e.g., graphs, charts] that can break-up the reader’s experience with the narrative flow of the study. Analyzing articles helps students practice comprehending these elements of writing, critiquing the arguments being made, reflecting upon the significance of the research, and how it relates to building new knowledge and understanding or applying new approaches to practice. Comprehending scholarly writing also involves thinking critically about where you fit within the overall dialogue among scholars concerning the research problem, finding possible gaps in the research that require further analysis, or identifying where the author(s) has failed to examine fully any specific elements of the study.
In addition, journal article analysis assignments are used by professors to strengthen discipline-specific information literacy skills, either alone or in relation to other tasks, such as, giving a class presentation or participating in a group project. These benefits can include the ability to:
- Effectively paraphrase text, which leads to a more thorough understanding of the overall study;
- Identify and describe strengths and weaknesses of the study and their implications;
- Relate the article to other course readings and in relation to particular research concepts or ideas discussed during class;
- Think critically about the research and summarize complex ideas contained within;
- Plan, organize, and write an effective inquiry-based paper that investigates a research study, evaluates evidence, expounds on the author’s main ideas, and presents an argument concerning the significance and impact of the research in a clear and concise manner;
- Model the type of source summary and critique you should do for any college-level research paper; and,
- Increase interest and engagement with the research problem of the study as well as with the discipline.
Kershaw, Trina C., Jennifer Fugate, and Aminda J. O'Hare. "Teaching Undergraduates to Understand Published Research through Structured Practice in Identifying Key Research Concepts." Scholarship of Teaching and Learning in Psychology . Advance online publication, 2020; Franco, Josue. “Introducing the Analysis of Journal Articles.” Prepared for presentation at the American Political Science Association’s 2020 Teaching and Learning Conference, February 7-9, 2020, Albuquerque, New Mexico; Sego, Sandra A. and Anne E. Stuart. "Learning to Read Empirical Articles in General Psychology." Teaching of Psychology 43 (2016): 38-42; Woodward-Kron, Robyn. "Critical Analysis and the Journal Article Review Assignment." Prospect 18 (August 2003): 20-36; MacMillan, Margy and Allison MacKenzie. "Strategies for Integrating Information Literacy and Academic Literacy: Helping Undergraduate Students make the most of Scholarly Articles." Library Management 33 (2012): 525-535; Kershaw, Trina C., Jordan P. Lippman, and Jennifer Fugate. "Practice Makes Proficient: Teaching Undergraduate Students to Understand Published Research." Instructional Science 46 (2018): 921-946.
Structure and Organization
A journal article analysis paper should be written in paragraph format and include an instruction to the study, your analysis of the research, and a conclusion that provides an overall assessment of the author's work, along with an explanation of what you believe is the study's overall impact and significance. Unless the purpose of the assignment is to examine foundational studies published many years ago, you should select articles that have been published relatively recently [e.g., within the past few years].
Since the research has been completed, reference to the study in your paper should be written in the past tense, with your analysis stated in the present tense [e.g., “The author portrayed access to health care services in rural areas as primarily a problem of having reliable transportation. However, I believe the author is overgeneralizing this issue because...”].
Introduction Section
The first section of a journal analysis paper should describe the topic of the article and highlight the author’s main points. This includes describing the research problem and theoretical framework, the rationale for the research, the methods of data gathering and analysis, the key findings, and the author’s final conclusions and recommendations. The narrative should focus on the act of describing rather than analyzing. Think of the introduction as a more comprehensive and detailed descriptive abstract of the study.
Possible questions to help guide your writing of the introduction section may include:
- Who are the authors and what credentials do they hold that contributes to the validity of the study?
- What was the research problem being investigated?
- What type of research design was used to investigate the research problem?
- What theoretical idea(s) and/or research questions were used to address the problem?
- What was the source of the data or information used as evidence for analysis?
- What methods were applied to investigate this evidence?
- What were the author's overall conclusions and key findings?
Critical Analysis Section
The second section of a journal analysis paper should describe the strengths and weaknesses of the study and analyze its significance and impact. This section is where you shift the narrative from describing to analyzing. Think critically about the research in relation to other course readings, what has been discussed in class, or based on your own life experiences. If you are struggling to identify any weaknesses, explain why you believe this to be true. However, no study is perfect, regardless of how laudable its design may be. Given this, think about the repercussions of the choices made by the author(s) and how you might have conducted the study differently. Examples can include contemplating the choice of what sources were included or excluded in support of examining the research problem, the choice of the method used to analyze the data, or the choice to highlight specific recommended courses of action and/or implications for practice over others. Another strategy is to place yourself within the research study itself by thinking reflectively about what may be missing if you had been a participant in the study or if the recommended courses of action specifically targeted you or your community.
Possible questions to help guide your writing of the analysis section may include:
Introduction
- Did the author clearly state the problem being investigated?
- What was your reaction to and perspective on the research problem?
- Was the study’s objective clearly stated? Did the author clearly explain why the study was necessary?
- How well did the introduction frame the scope of the study?
- Did the introduction conclude with a clear purpose statement?
Literature Review
- Did the literature review lay a foundation for understanding the significance of the research problem?
- Did the literature review provide enough background information to understand the problem in relation to relevant contexts [e.g., historical, economic, social, cultural, etc.].
- Did literature review effectively place the study within the domain of prior research? Is anything missing?
- Was the literature review organized by conceptual categories or did the author simply list and describe sources?
- Did the author accurately explain how the data or information were collected?
- Was the data used sufficient in supporting the study of the research problem?
- Was there another methodological approach that could have been more illuminating?
- Give your overall evaluation of the methods used in this article. How much trust would you put in generating relevant findings?
Results and Discussion
- Were the results clearly presented?
- Did you feel that the results support the theoretical and interpretive claims of the author? Why?
- What did the author(s) do especially well in describing or analyzing their results?
- Was the author's evaluation of the findings clearly stated?
- How well did the discussion of the results relate to what is already known about the research problem?
- Was the discussion of the results free of repetition and redundancies?
- What interpretations did the authors make that you think are in incomplete, unwarranted, or overstated?
- Did the conclusion effectively capture the main points of study?
- Did the conclusion address the research questions posed? Do they seem reasonable?
- Were the author’s conclusions consistent with the evidence and arguments presented?
- Has the author explained how the research added new knowledge or understanding?
Overall Writing Style
- If the article included tables, figures, or other non-textual elements, did they contribute to understanding the study?
- Were ideas developed and related in a logical sequence?
- Were transitions between sections of the article smooth and easy to follow?
Overall Evaluation Section
The final section of a journal analysis paper should bring your thoughts together into a coherent assessment of the value of the research study . This section is where the narrative flow transitions from analyzing specific elements of the article to critically evaluating the overall study. Explain what you view as the significance of the research in relation to the overall course content and any relevant discussions that occurred during class. Think about how the article contributes to understanding the overall research problem, how it fits within existing literature on the topic, how it relates to the course, and what it means to you as a student researcher. In some cases, your professor will also ask you to describe your experiences writing the journal article analysis paper as part of a reflective learning exercise.
Possible questions to help guide your writing of the conclusion and evaluation section may include:
- Was the structure of the article clear and well organized?
- Was the topic of current or enduring interest to you?
- What were the main weaknesses of the article? [this does not refer to limitations stated by the author, but what you believe are potential flaws]
- Was any of the information in the article unclear or ambiguous?
- What did you learn from the research? If nothing stood out to you, explain why.
- Assess the originality of the research. Did you believe it contributed new understanding of the research problem?
- Were you persuaded by the author’s arguments?
- If the author made any final recommendations, will they be impactful if applied to practice?
- In what ways could future research build off of this study?
- What implications does the study have for daily life?
- Was the use of non-textual elements, footnotes or endnotes, and/or appendices helpful in understanding the research?
- What lingering questions do you have after analyzing the article?
NOTE: Avoid using quotes. One of the main purposes of writing an article analysis paper is to learn how to effectively paraphrase and use your own words to summarize a scholarly research study and to explain what the research means to you. Using and citing a direct quote from the article should only be done to help emphasize a key point or to underscore an important concept or idea.
Business: The Article Analysis . Fred Meijer Center for Writing, Grand Valley State University; Bachiochi, Peter et al. "Using Empirical Article Analysis to Assess Research Methods Courses." Teaching of Psychology 38 (2011): 5-9; Brosowsky, Nicholaus P. et al. “Teaching Undergraduate Students to Read Empirical Articles: An Evaluation and Revision of the QALMRI Method.” PsyArXi Preprints , 2020; Holster, Kristin. “Article Evaluation Assignment”. TRAILS: Teaching Resources and Innovations Library for Sociology . Washington DC: American Sociological Association, 2016; Kershaw, Trina C., Jennifer Fugate, and Aminda J. O'Hare. "Teaching Undergraduates to Understand Published Research through Structured Practice in Identifying Key Research Concepts." Scholarship of Teaching and Learning in Psychology . Advance online publication, 2020; Franco, Josue. “Introducing the Analysis of Journal Articles.” Prepared for presentation at the American Political Science Association’s 2020 Teaching and Learning Conference, February 7-9, 2020, Albuquerque, New Mexico; Reviewer's Guide . SAGE Reviewer Gateway, SAGE Journals; Sego, Sandra A. and Anne E. Stuart. "Learning to Read Empirical Articles in General Psychology." Teaching of Psychology 43 (2016): 38-42; Kershaw, Trina C., Jordan P. Lippman, and Jennifer Fugate. "Practice Makes Proficient: Teaching Undergraduate Students to Understand Published Research." Instructional Science 46 (2018): 921-946; Gyuris, Emma, and Laura Castell. "To Tell Them or Show Them? How to Improve Science Students’ Skills of Critical Reading." International Journal of Innovation in Science and Mathematics Education 21 (2013): 70-80; Woodward-Kron, Robyn. "Critical Analysis and the Journal Article Review Assignment." Prospect 18 (August 2003): 20-36; MacMillan, Margy and Allison MacKenzie. "Strategies for Integrating Information Literacy and Academic Literacy: Helping Undergraduate Students Make the Most of Scholarly Articles." Library Management 33 (2012): 525-535.
Writing Tip
Not All Scholarly Journal Articles Can Be Critically Analyzed
There are a variety of articles published in scholarly journals that do not fit within the guidelines of an article analysis assignment. This is because the work cannot be empirically examined or it does not generate new knowledge in a way which can be critically analyzed.
If you are required to locate a research study on your own, avoid selecting these types of journal articles:
- Theoretical essays which discuss concepts, assumptions, and propositions, but report no empirical research;
- Statistical or methodological papers that may analyze data, but the bulk of the work is devoted to refining a new measurement, statistical technique, or modeling procedure;
- Articles that review, analyze, critique, and synthesize prior research, but do not report any original research;
- Brief essays devoted to research methods and findings;
- Articles written by scholars in popular magazines or industry trade journals;
- Pre-print articles that have been posted online, but may undergo further editing and revision by the journal's editorial staff before final publication; and
- Academic commentary that discusses research trends or emerging concepts and ideas, but does not contain citations to sources.
Journal Analysis Assignment - Myers . Writing@CSU, Colorado State University; Franco, Josue. “Introducing the Analysis of Journal Articles.” Prepared for presentation at the American Political Science Association’s 2020 Teaching and Learning Conference, February 7-9, 2020, Albuquerque, New Mexico; Woodward-Kron, Robyn. "Critical Analysis and the Journal Article Review Assignment." Prospect 18 (August 2003): 20-36.
- << Previous: Annotated Bibliography
- Next: Giving an Oral Presentation >>
- Last Updated: Mar 6, 2024 1:00 PM
- URL: https://libguides.usc.edu/writingguide/assignments

Provide details on what you need help with along with a budget and time limit. Questions are posted anonymously and can be made 100% private.

Studypool matches you to the best tutor to help you with your question. Our tutors are highly qualified and vetted.

Your matched tutor provides personalized help according to your question details. Payment is made only after you have completed your 1-on-1 session and are satisfied with your session.

- Homework Q&A
- Become a Tutor
All Subjects
Mathematics
Programming
Health & Medical
Engineering
Computer Science
Foreign Languages
Access over 20 million homework & study documents
Nr 449 unit 5 required uniform assignment rua 2 analyzing published research.
Sign up to view the full document!

24/7 Homework Help
Stuck on a homework question? Our verified tutors can answer all questions, from basic math to advanced rocket science !

Similar Documents
working on a homework question?
Studypool is powered by Microtutoring TM
Copyright © 2024. Studypool Inc.
Studypool is not sponsored or endorsed by any college or university.
Ongoing Conversations

Access over 20 million homework documents through the notebank
Get on-demand Q&A homework help from verified tutors
Read 1000s of rich book guides covering popular titles

Sign up with Google
Sign up with Facebook
Already have an account? Login
Login with Google
Login with Facebook
Don't have an account? Sign Up
NR 449 Unit 5 Required Uniform Assignment (RUA) 2; Analyzi... - $14.45 Add to Cart
NR 449 Unit 5 Required Uniform Assignment (RUA) 2; Analyzing Published Research.
Preview 1 out of 7 pages

Generating Your Document
Purchase the document to get the full access instantly
- 3 Million+ Study Documents
- 100% Money Back Guarantee
- Immediate Download Access

Report Copyright Violation
Exam Details
Add To Cart
Add To Wishlist
- Trusted by 800,000+ Students
- 24/7 Money Back Guarantee
- Download is directly available
Specifications
Institution South University
Study NURSING
Course NR 449 Unit 5 Required Uniform Assignment (RUA) 2; Analyzing Published Research.
Language English
Subject Health Care
Updated On Oct 09,2021
Number of Pages 7
Written 2020-2021
Seller Details

2978 documents uploaded
87 documents sold
Recommended documents

NR 449 Unit 5 Required Un...

NAPSRx Final Exam Solutio...

Focused Exam: Abdominal P...

HLTH 501 FINAL EXAM | Com...

BUS 660 Forecasting Case ...
What students are saying about us.
Docmerit is a great platform to get and share study resources, especially the resource contributed by past students.

Northwestern University Karen
I find Docmerit to be authentic, easy to use and a community with quality notes and study tips. Now is my chance to help others.

University Of Arizona Anna Maria
One of the most useful resource available is 24/7 access to study guides and notes. It helped me a lot to clear my final semester exams.

Devry University David Smith
Docmerit is super useful, because you study and make money at the same time! You even benefit from summaries made a couple of years ago.

Liberty University Mike T
- Follow us on
- Copyright © 2024 | All rights reserved
Register a free account

Tutorspedia
Place order.
File Upload : File Upload :

Reaserch-QA65 Online Services
Required uniform assignment: analyzing published research.
The purpose of this paper is to interpret the two articles identified as most important to the group topic.
Course Outcomes
This assignment enables the student to meet the following course outcomes CO 2: Apply research principles to the interpretation of the content of published research studies. CO 4: Evaluate published nursing research for credibility and clinical significance related to evidence- based practice.
Total Points Possible :200 Points Requirements
How it works, how it works .
Step 1:- Click on Submit your Assignment here or shown in top menu of every page and fill the quotation form with all the details. In the comment section, please mention product code mentioned in end of every Q&A Page . You can also send us your details through our email id [email protected] with product code in the email body. Product code is essential to locate your questions so please mentioned that in your email or submit your quotes form comment section.
Step 2:- While filling submit your quotes form please fill all details like deadline date, expected budget, topic , your comments in addition to product code . The date is asked to provide deadline.
Step 3:- Once we received your assignments through submit your quotes form or email, we will review the Questions and notify our price through our email id. Kindly ensure that our email id [email protected] must not go into your spam folders. We request you to provide your expected budget as it will help us in negotiating with our experts.
Step 4:- Once you agreed with our price, kindly pay by clicking on Pay Now and please ensure that while entering your credit card details for making payment, it must be done correctly and address should be your credit card billing address. You can also request for invoice to our live chat representatives.
Step 5:- Once we received the payment we will notify through our email and will deliver the Q&A solution through mail as per agreed upon deadline.
Step 6:- You can also call us in our phone no. as given in the top of the home page or chat with our customer service representatives by clicking on chat now given in the bottom right corner.
Features for Assignment Help
Related services.
- Accounting Homework Help
- Business Law Homework Help
- Management Homework Help
- Finance Tutor Help
- Finance Homework Help
- Geology Homework Help
- HR Management Homework Help
- Humanities Homework Help
- Information Management Help
- Linguistics Homework Help
- Managerial Accounting Help
- Mathematics Tutor Help
- Nursing Homework Help
- Operations Management Help
- Project Management Homework
- Statistics Tutor Help
- Essay Writing Services
- Writing Services Australia
- Finance Online Exam Help
- Writing Services In UK
2. Description of Findings
a. Summarize basics in the Matrix Table as found in Assignment Documents in e-College. b. Describe i. Concepts ii. Methods used iii. Participants iv. Instruments including reliability and validity v. Answer to “Purpose” question vi. Identify next step for group
3. Conclusion of paper
a. Correct grammar and spelling b. Use of headings for each section NR449 Evidence Based Practice c. Use of APA format (sixth edition) d. Page length: three pages
Preparing the Paper
1. Please make sure you do not duplicate articles within your group. 2. Paper should include a title page and a reference page.
Product code:Reaserch-QA65
Looking for Best Reaserch-QA65, please click here
Reimagining Design with Nature: ecological urbanism in Moscow
- Reflective Essay
- Published: 10 September 2019
- Volume 1 , pages 233–247, ( 2019 )
Cite this article
- Brian Mark Evans ORCID: orcid.org/0000-0003-1420-1682 1
976 Accesses
2 Citations
Explore all metrics
The twenty-first century is the era when populations of cities will exceed rural communities for the first time in human history. The population growth of cities in many countries, including those in transition from planned to market economies, is putting considerable strain on ecological and natural resources. This paper examines four central issues: (a) the challenges and opportunities presented through working in jurisdictions where there are no official or established methods in place to guide regional, ecological and landscape planning and design; (b) the experience of the author’s practice—Gillespies LLP—in addressing these challenges using techniques and methods inspired by McHarg in Design with Nature in the Russian Federation in the first decade of the twenty-first century; (c) the augmentation of methods derived from Design with Nature in reference to innovations in technology since its publication and the contribution that the art of landscape painters can make to landscape analysis and interpretation; and (d) the application of this experience to the international competition and colloquium for the expansion of Moscow. The text concludes with a comment on how the application of this learning and methodological development to landscape and ecological planning and design was judged to be a central tenant of the winning design. Finally, a concluding section reflects on lessons learned and conclusions drawn.
This is a preview of subscription content, log in via an institution to check access.
Access this article
Price includes VAT (Russian Federation)
Instant access to the full article PDF.
Rent this article via DeepDyve
Institutional subscriptions
Similar content being viewed by others

The politics of designing with nature: reflections from New Orleans and Dhaka
Zachary Lamb

Acknowledgements
The landscape team from Gillespies Glasgow Studio (Steve Nelson, Graeme Pert, Joanne Walker, Rory Wilson and Chris Swan) led by the author and all our collaborators in the Capital Cities Planning Group.
Author information
Authors and affiliations.
Mackintosh School of Architecture, The Glasgow School of Art, 167 Renfrew Street, Glasgow, G3 6BY, UK
Brian Mark Evans
You can also search for this author in PubMed Google Scholar
Corresponding author
Correspondence to Brian Mark Evans .
Rights and permissions
Reprints and permissions

About this article
Evans, B.M. Reimagining Design with Nature: ecological urbanism in Moscow. Socio Ecol Pract Res 1 , 233–247 (2019). https://doi.org/10.1007/s42532-019-00031-5
Download citation
Received : 17 March 2019
Accepted : 13 August 2019
Published : 10 September 2019
Issue Date : October 2019
DOI : https://doi.org/10.1007/s42532-019-00031-5
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Design With Nature
- Find a journal
- Publish with us
- Track your research
Thank you for visiting nature.com. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser (or turn off compatibility mode in Internet Explorer). In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript.
- View all journals
- My Account Login
- Explore content
- About the journal
- Publish with us
- Sign up for alerts
- Open access
- Published: 15 March 2024
Metagenomic insights into the wastewater resistome before and after purification at large‑scale wastewater treatment plants in the Moscow city
- Shahjahon Begmatov 1 ,
- Alexey V. Beletsky 1 ,
- Alexander G. Dorofeev 2 ,
- Nikolai V. Pimenov 2 ,
- Andrey V. Mardanov 1 &
- Nikolai V. Ravin 1
Scientific Reports volume 14 , Article number: 6349 ( 2024 ) Cite this article
492 Accesses
Metrics details
- Antimicrobials
- Microbial communities
- Industrial microbiology
Wastewater treatment plants (WWTPs) are considered to be hotspots for the spread of antibiotic resistance genes (ARGs). We performed a metagenomic analysis of the raw wastewater, activated sludge and treated wastewater from two large WWTPs responsible for the treatment of urban wastewater in Moscow, Russia. In untreated wastewater, several hundred ARGs that could confer resistance to most commonly used classes of antibiotics were found. WWTPs employed a nitrification/denitrification or an anaerobic/anoxic/oxic process and enabled efficient removal of organic matter, nitrogen and phosphorus, as well as fecal microbiota. The resistome constituted about 0.05% of the whole metagenome, and after water treatment its share decreased by 3–4 times. The resistomes were dominated by ARGs encoding resistance to beta-lactams, macrolides, aminoglycosides, tetracyclines, quaternary ammonium compounds, and sulfonamides. ARGs for macrolides and tetracyclines were removed more efficiently than beta-lactamases, especially ampC , the most abundant ARG in the treated effluent. The removal efficiency of particular ARGs was impacted by the treatment technology. Metagenome-assembled genomes of multidrug-resistant strains were assembled both for the influent and the treated effluent. Ccomparison of resistomes from WWTPs in Moscow and around the world suggested that the abundance and content of ARGs depend on social, economic, medical, and environmental factors.
Similar content being viewed by others

Metagenomic analysis of an urban resistome before and after wastewater treatment
Felipe Lira, Ivone Vaz-Moreira, … José Luis Martínez
Mobile resistome of microbial communities and antimicrobial residues from drinking water supply systems in Rio de Janeiro, Brazil
Kayo Bianco, Beatriz Oliveira de Farias, … Maysa Mandetta Clementino
Differential effects of wastewater treatment plant effluents on the antibiotic resistomes of diverse river habitats
Jangwoo Lee, Feng Ju, … Helmut Bürgmann
Introduction
The spread of antimicrobial resistance (AMR) in the environmental microbiome has become one of the frequently noted problems in recent years, along with global climate change, food security and other technological challenges. Numerous studies show that from year to year, in addition to increasing the cost of hospitalization and treatment of patients infected with multidrug-resistant bacteria, the number of deaths of such patients is growing 1 , 2 . Understanding the mechanisms underlying the emergence, selection and dissemination of AMR, and antibiotic resistance genes (ARGs), is required for the development of sustainable strategies to control and minimize this threat. The dissemination of antibiotic resistant bacteria (ARB) and ARGs occurs differently and this process is more active in urban territories rather than in rural ones. The rate of spread of ARGs and ARB in urban areas is obviously determined by the high population density and, as a rule, wastewater which flows from these areas contains both ARG and ARB. Most antibiotics used in medicine, agriculture and the food industry, as well as resistant bacteria, end up in wastewater. Wastewater treatment plants (WWTPs) therefore could provide a comprehensive overview of ARG abundance, diversity and genomic backgrounds in particular region 3 . Moreover, wastewater and WWTPs are places where ARGs and ARB are particularly abundant and are often considered “hotspots” for the formation of strains with multiple resistance and one of the main sources of the spread of AMR in the environment 4 .
Despite numerous studies on the role of WWTPs in resistome diversity and dissemination, each new study is, in terms of time and geography, unique, as many urban areas and countries have not yet been studied. In addition, some studies are dedicated to explore only one component of the wastewater treatment system, such as wastewater, activated sludge or treated effluent, and there is a lack of research that would give a comprehensive view of the diversity and change in the composition of the resistome at different stages of water cleaning, from wastewater to treated effluent, released into the environment.
Usually, wastewater treatment in large facilities takes place in three stages. The first stage includes physical methods of water cleaning, the second stage is microbiological treatment in bioreactors with activated sludge (AS), and the third stage is the final treatment of water and its disinfection. At the second stage, than could be performed using several technologies, microorganisms of AS are used to remove organic matter, ammonium, and (in more complex processes) phosphorus 5 . At this stage, the removal of microorganisms present in the wastewater, including ARB, occurs due to their adsorption on AS particles, which are removed along with excess AS. The efficiency of this process differs for various bacteria and depends on the purification technology used. Therefore, purification technologies directly affect the removal of particular ARGs and ARB, however, this issue was poorly studied 6 .
ARGs representing all known resistance mechanisms have been found in WWTP environments 7 . ARGs for beta-lactams, macrolides, quinolones, tetracyclines, sulfonamides, trimethoprim, and multidrug efflux pump genes have been found in the incoming wastewater, AS, and treated effluent in various countries 7 , 8 . Recently, Munk and coauthors (2022) using metagenomics methods characterized resistomes of 757 urban wastewater samples from 243 cities in 101 countries covering 7 major geographical regions. They reported regional patterns in wastewater resistomes that differed between subsets corresponding to drug classes and were partly driven by taxonomic variation 3 . Although this study did not analyzes the composition of the wastewater resistome after treatment, there are numerous evidences that the prevalence of ARB and ARG in rivers may increase downstream from the point of discharge of treated wastewater into them 9 , 10 . In a study of WWTPs in Germany, 123 types of clinically significant antibiotic resistance genes were found in treated wastewater discharged into water bodies 11 . An analysis of the presence of 30 ARGs at different stages of wastewater treatment at WWTPs in Northern China showed that the content of most ARGs in the treated effluent was lower compared to the influent entering the treatment, although an increase in the abundance of some ARGs and their release into the environment was also observed 12 . A metagenomic analysis of WWTP in Hong Kong revealed seasonal changes in the content of several types of ARG and its decrease in the treated effluent 13 , 14 . Most ARGs were reduced by more than 98% in the treated effluent compared to the wastewater entering the treatment 14 . Some other studies have also reported a decrease in ARGs after wastewater treatment 15 , 16 , 17 . However, in other studies, no changes in the ARG content or even an increase were observed 17 , 18 , 19 . Although there are numerous studies of resistomes in WWTP-related environments the distribution of samples was geographically biased and covered mostly North America, Western Europe, Eastern Asia (mostly China), Australasia, and few places in South America/Caribbean and Sub-Saharan Africa 3 .
In order to expand the geographical coverage and our knowledge about global resistome abundance and diversity, we analyzed resistomes of wastewater before and after treatment at large-scale WWTPs in the city of Moscow (Russia). Although Moscow WWTPs are among the largest in the world and may play an important role in the spread of antibiotic resistance, the resistomes of municipal wastewaters in Moscow have not previously been studied by modern molecular genetic methods. Previously we performed 16S rRNA gene profiling of AS microbial communities at large-scale WWTPs responsible for the treatment of municipal wastewater ion Moscow 5 . Comparison of microbial communities of AS samples from WWTPs in Moscow and worldwide revealed that Moscow samples clustered together indicating the importance of influent characteristics, related to local social and environmental factors, for wastewater microbiomes 5 . For example, due to the relatively low cost of water for household consumption, wastewater in Moscow has a relatively low content of organic matter. Apparently the presence of ARB and ARGs in communal wastewater depends on the frequency of antibiotic use and the range of drugs used. These factors differ in different countries and cities. Therefore, the characterization of the resistome and the role of Moscow WWTPs in the dispersion of ARGs is an important goal. Of particular interest is also the assessment of the impact of wastewater treatment technology on the composition of the resistome and the degree of ARG removal.
Here we present the first metagenomic overview of the composition of resistome of influent wastewater, AS and treated effluent released into the environment at two Moscow WWTPs employing different treatment technologies.
Characteristics of WWTPs and water chemistry
The Lyuberetskiy WWTP complex of JSC “Mosvodokanal” carry out the treatment of wastewater in the city of Moscow with a capacity of about 2 million m 3 per day. This complex consists of several wastewater treatment units (hereafter referred to as WWTPs). They purify the same inflow wastewater but otherwise are independent installations between which there is no transfer and mixing of AS. Two WWTPs implementing different technologies for wastewater treatment were chosen as the objects of study. The first one (LOS) is operated using anaerobic/anoxic/oxic process, also known as the University of Cape Town (UCT) technology. There the sludge mixture first enters the anaerobic zone, where phosphate-accumulating microorganisms (PAO) consume easily degradable organics, then to the anoxic zone, where denitrification and accumulation of phosphates by denitrifying PAO occur, and finally to the aerobic zone, where organic matter and ammonium are oxidized while PAO accumulate large quantities of polyphosphate. The second WWTP (NLOS2) uses a simpler nitrification–denitrification technology (N-DN). In the aerobic zones organics and ammonium are oxidized, while in the anoxic zone nitrate is reduced to gaseous nitrogen. This treatment technology removes organic matter and nitrogen, but was not specially aimed to remove phosphates. The production capacity of LOS is approximately 2 times more than that of NLOS2; there are no other important differences between these WWTPs besides treatment technology.
Sampling and chemical analysis
Wastewater and AS samples were collected in September 2022 and kindly provided by “Mosvodokanal” JSC. The temperature of water samples was about 24 °C. Samples of AS from bioreactors of two WWTPs were taken in 50 ml Falcon tubes (BD Biosciences). Wastewater samples (influent and effluents from two WWTPs) were taken in 5 L plastic bottles. The cells were collected by centrifugation at 3000 g for 20 min at 4 °C.
Wastewater quality values, namely, biochemical oxygen demand (five days incubation) (BOD 5 ), chemical oxygen demand (COD), total suspended solids (TSS), sludge volume index (SVI), ammonium nitrogen (N-NH 4 ), nitrate nitrogen (N-NO 3 ), nitrite nitrogen (N-NO 2 ) and phosphorus (P-PO 4 ) in the influent and effluents of two WWTPs were measured by the specialized laboratory “MSULab” according to the Federal inspection of environmental management’s protocols for chemical analyses of water.
DNA isolation, 16S rRNA gene sequencing and analysis
Total genomic DNA was isolated using a Power Soil DNA isolation kit (Qiagen, Germany). DNA for each sample was isolated in four parallel replicates, which were then pooled. PCR amplification of 16S rRNA gene fragments comprising the V3–V4 variable regions was performed using the universal primers 341F (5′-CCTAYG GGDBGCWSCAG) and 806R (5′-GGA CTA CNVGGG THTCTAAT) 20 . The obtained PCR fragments were bar-coded and sequenced on Illumina MiSeq (2 × 300 nt reads). Pairwise overlapping reads were merged using FLASH v.1.2.11 21 . All sequences were clustered into operational taxonomic units (OTUs) at 97% identity using the USEARCH v.11 program 22 . Low quality reads were removed prior to clustering, chimeric sequences and singletons were removed during clustering by the USEARCH algorithms. To calculate OTU abundances, all reads obtained for a given sample were mapped to OTU sequences at a 97% global identity threshold by USEARCH. The taxonomic assignment of OTUs was performed by searching against the SILVA v.138 rRNA sequence database using the VSEARCH v. 2.14.1 algorithm 23 .
The diversity indices at a 97% OTU cut-off level were calculated using USEARCH v.11 22 . To avoid sequencing depth bias, the numbers of reads for each sample were randomly sub-sampled to the size of the smallest set.
Sequencing of metagenomic DNA, contigs assembly and binning of MAGs
Metagenomic DNA was sequenced using the Illumina HiSeq2500 platform according to the manufacturer’s instructions (Illumina Inc., San Diego, CA, USA). The sequencing of a paired-end (2 × 150 bp) NEBNext Ultra II DNA Library prep kit (NEB) generated from 145 to 257 million read pairs per sample. Adapter removal and trimming of low-quality sequences (Q < 30) were performed using Cutadapt v.3.4 24 and Sickle v.1.33 ( https://github.com/najoshi/sickle ), respectively. The resulting Illumina reads were de novo assembled into contigs using SPAdes v.3.15.4 in metagenomic mode 25 .
The obtained contigs were binned into metagenome-assembled genomes (MAGs) using 3 different programs: MetaBAT v.2.2.15 26 , MaxBin v.2.2.7 27 and CONCOCT v.1.1.0 28 . The results of the three binning programs were merged into an optimized set of MAGs using DAS Tool v.1.1.4 29 . The completeness of the MAGs and their possible contamination (redundancy) were estimated using CheckM v.1.1.3 30 with lineage-specific marker genes. The assembled MAGs were taxonomically classified using the Genome Taxonomy Database Toolkit (GTDB-Tk) v.2.0.0 31 and Genome Taxonomy database (GTDB) 32 .
ARG identification
Open reading frames (ORFs) were predicted in assembled contigs using Prodigal v.2.6.3 33 . ARGs were predicted using the NCBI AMRFinderPlus v.3.11.4 ( https://github.com/ncbi/amr/wiki ) command line tool and its associated database 34 . The predicted protein sequences of all ORFs were analyzed in this tool with parameter “-p”.
Efficiency of wastewater treatment
Two wastewater treatment technologies were used in the investigated WWTPs,—nitrification/denitrification at NLOS2 and more advanced anaerobic/anoxic/oxic UCT process at LOS. LOS removed more than 99.5% of organic matter (according to the BOD5 data) and more than 99.9% of ammonium while the performance of NLOS2 was poorer (Table 1 ). Particularly noticeable differences were observed in nitrate and nitrite concentrations in the effluents suggesting the lower efficiency of denitrification in the NLOS2. Interestingly, although the NLOS2 unit was not designed to remove phosphorus, the concentration of phosphates in the treated effluent at this WWTP is only slightly higher than at LOS. The treated influent at LOS contains fewer solids consistently with lower SVI. Overall, the technology used at LOS plant is more efficient.
Microbiomes of the influent wastewater, activated sludge and treated effluent
The 16S rRNA gene profiling of microbial communities revealed 1013 species-level OTUs (97% identity) in the influent and 1.2–1.7 times more OTUs in the AS and treated effluent samples (Supplemental Table S1 ). The Shannon diversity indices correlated with the number of detected OTUs and increased in the series “influent” – “activated sludge” – “effluent” at each WWTP (Supplemental Table S2 ).
Analysis of the microbiome of wastewater supplied for biological treatment showed that that the most numerous phyla in the microbial community were Firmicutes (28.4% of all 16S rRNA gene sequences), Campylobacterota (28.0%), Proteobacteria (20.9%), and Bacteroidota (10.5%) (Fig. 1 ). These were mainly representatives of the fecal microbiota, which are often found in wastewater. The phylum Firmicutes was dominated by Streptococcaceae (9.7%, mostly S treptococcus sp.), Lachnospiraceae (5.9%), Ruminococcaceae (3.0%), Carnobacteriaceae (1.7%), Peptostreptococcaceae (1.6%) and Veillonellaceae (1.4%). Most of Campylobacterota belonged to the family Arcobacteraceae (26.8%) of the genera Arcobacter (19.9%), Pseudarcobacter (2.5%) and uncultured lineage (4.3%), as well as by sulfur-oxidizing Sulfurospirillum (1.0%). Among the Proteobacteria the most abundant genera were Acinetobacter (7.8%) , Aeromonas (1.8%) and Pseudomonas (1.1%). Most of the identified Bacteroidota were typical fecal contaminants such as members of the genera Bacteroides (2.6%), Macellibacteroides (1.5%), Prevotella (1.4%), and Cloacibacterium (1.2%).
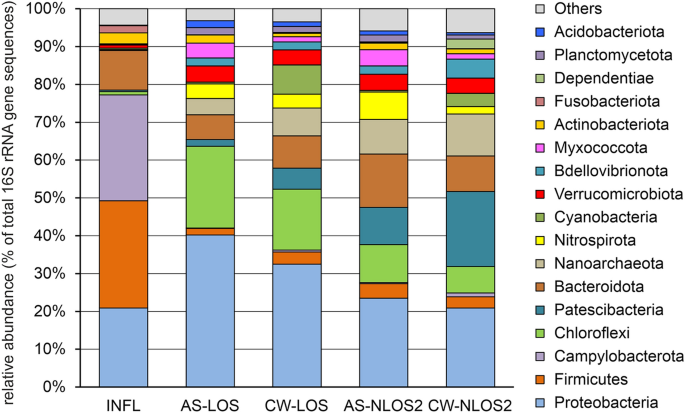
Microbial community composition in the influent, AS and treated effluent samples according to 16S rRNA gene profiling. The composition is displayed at the phylum level. INFL, influent wastewater; AS-LOS, AS at LOS plant; CW-LOS, treated effluent at LOS plant; AS-NLOS2, AS at NLOS2 plant; CW-NLOS2, treated effluent at NLOS2 plant.
Activated sludge of WWTP bioreactors is a complex microbial community consisting of physiologically and phylogenetically heterogeneous groups of microorganisms involved in the removal of major contaminants from wastewater. The composition of AS microbiomes was very different from the microbiome of incoming wastewater (Fig. 1 ). The phyla Campylobacterota (less than 0.5%) and Firmicutes (2–4%) were much less abundant in AS microbiomes. Proteobacteria was the dominant group in the microbiomes of AS (23–40%), but its composition differed from the microbiome of influent wastewater: instead of the fecal microflora (Enterobacterales and others) the AS community harbored lineages involved in the purification processes ( Competibacteraceae , Rhodocyclaceae , Nitrosomonadaceae , etc.). Likewise, Bacteroidota were among the most numerous phyla in AS microbiomes at both LOS (6.5%) and NLOS2 (14.1%), but instead of Bacteroidales mostly comprised Chitinophagales and Sphingobacteriales typical for AS communities. The numerous groups of AS community also included Chloroflexi (22% and 10% in LOS and NLOS2, respectively), Patescibacteria (1.8% and 9.9%), Nanoarchaeota (4.3% and 9.1%), Nitrospirota (3.9% and 7.3%), Verrucomicrobiota and Myxococcota (about 4% in both WWTPs). Bacteria that play an important role in the removal of nitrogen ( Nitrospira and Nitrosomonas ) and phosphorus ( Dechloromonas ), as well as glycogen-accumulating Ca . Competibacter, have been found in large numbers. The abundance of these functional groups is consistent with the high efficiency of nitrogen and phosphorus removal.
The main source of microorganisms in treated effluent is the AS, from which they are washed out; bacteria from the influent water may also be present in minor amounts. Therefore, as expected, the microbiome composition of treated wastewater was similar to that of activated sludge. Consistently, compositions of microbiomes of treated effluent were similar to that of AS samples. However, some differences were observed, in particular, the microbiomes of the treated effluent contained many Cyanobacteria (7.74% and 3.49% for LOS and NLOS2, respectively) which were found in minor amounts both in the influent water and in the ASs (< 0.5%). Probably, these light-dependent bacteria proliferate in the final clarifier and then can be easily washed out with the effluent.
Diversity of resistomes
The results of metagenomic analysis of incoming wastewater revealed 544 ARGs in the assembled contigs, classified into 33 AMR gene families (Table 2 and Supplemental Table S3 ). Among the most numerous were classes A, C, D and metallo- beta-lactamases, rifampin ADP-ribosyltransferase, Erm 23S ribosomal RNA methyltransferase, aminoglycoside nucleotidyl-, acetyl- and phospho-transferases, the ABC-F type ribosomal protection proteins, chloramphenicol acetyltransferase, trimethoprim-resistant dihydrofolate reductase, quaternary ammonium compound efflux SMR transporters, lincosamide nucleotidyltransferases, tetracycline efflux MFS transporters and tetracycline resistance ribosomal protection proteins (Table 2 ). These genes may enable antibiotic inactivation (373 genes), target protection (85 genes), efflux (44 genes) and target replacement (25 genes).
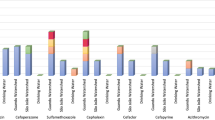
The abovementioned genes confer resistance to most of commonly used drugs: beta-lactams (198 genes), macrolides (74 genes), rifamycin (60 genes), aminoglycosides (51 genes), tetracycline (27 genes), phenicols (27 genes), diaminopyrimidines (19 genes), quaternary ammonium compounds (16 genes), glycopeptides (15 genes), lincosamide (13 genes), fosfomycine (12 genes) and drugs of 11 others classes (Fig. 2 ).
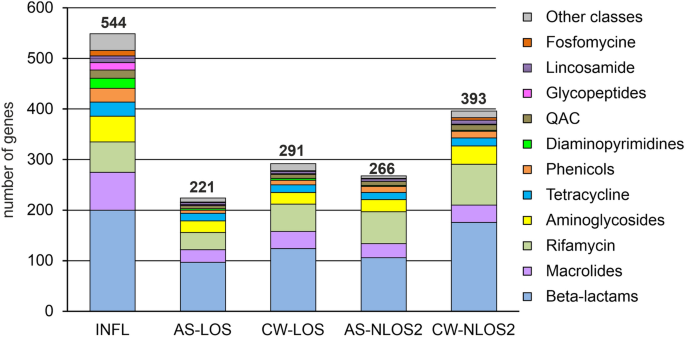
ARGs identified in wastewater and AS samples categorized by drug classes.
About twice less ARGs were identified in AS samples from both WWTPs. Like in the influent, beta-lactamases of classes A, D, and metallo-beta-lactamases were the most numerous, while only a few genes for class C enzymes were found (Table 2 ). Other families of ARGs, numerous in the influent, were also numerous in AS microbiomes. A notable difference between the resistomes of the AS samples is the greater number of rifampin-ADP-ribosyltransferase genes ( arr ) in NLOS2 compared to LOS (63 vs 33). The largest number of arr genes was assigned to Bacteroidota, and the lower relative abundance of this phylum in AS at LOS likely explains these differences. Like in the wastewater, resistance to beta-lactams, macrolides, rifamycin, aminoglycosides, and tetracyclines was the most common (Fig. 2 ). On the contrary, genes for some drug classes were underrepresented in AS resistomes, especially for diaminopyrimidines (3 and 2 genes for LOS and NLOS2, respectively) and glycopeptide antibiotics (2 and 0 genes).
The results of metagenomic analysis of treated effluent showed that the diversity of these resistomes was only slightly higher than that of the corresponding AS samples. This result was expected since the main source of microorganisms in the effluent is activated sludge, from which they are partially washed. However, resistomes of treated effluent at both WWTPs contains about twice more class A beta-lactamase genes than AS samples suggesting less efficient absorption of their host bacteria at AS particles (Table 2 ).
Quantitative analysis of antibiotic resistance genes of WWTP
The results described above provide information on the diversity of resistance genes, but not on their abundance in the metagenomes, which depends on the abundance of corresponding bacterial hosts. To quantify the shares of individual ARGs in the metagenome and resistome, the amounts of metagenomic reads mapped to the corresponding ARGs in contigs were determined. In total, the resistome accounted for about 0.05% of the metagenome of wastewater supplied for treatment, while the shares of resistomes in the metagenomes of AS and treated effluent samples were 0.02% and 0.014% at the LOS and NLOS2 WWTPs, respectively.
Quantitative analysis of the content of individual ARGs in metagenomes showed that the structure of the influent resistome was very different from that of AS and treated effluent. The relative content of ARGs accounting for more than 1% in at least one analyzed resistome is shown in Fig. 3 . The LOS and NLOS2 WWTPs differed significantly from each other, and the differences between the AS and effluent resistomes at each WWTP were much less pronounced.
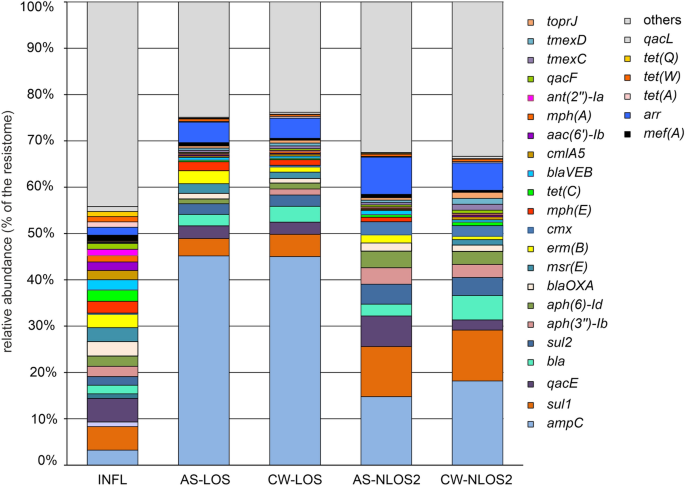
The relative abundancies of particular ARGs in the resistomes. Only ARGs with shares greater than 1% in at least one sample are shown, all other ARGs are shown as “others”.
The resistome of the influent was not only the most diverse, but also the most even in composition. The shares of none of the ARGs exceeded 5% of the resistome, and the 23 most common ARGs accounted for a half of the resistome. The most abundant ten ARGs were qacE, sul1, ampC, blaOXA, msr(E), erm(B), mph(E), tet(C), aph(3'')-Ib and aph(6)-Id, conferring resistance to antiseptics, sulfonamides, beta-lactams, macrolides, aminoglycosides (streptomycin), and tetracyclines.
AS and treated effluent at LOS plant was strongly dominated by a single AGR type, class C beta-lactamase ampC , accounting for about 45% of their resistomes. This gene was also the most abundant one in the resistomes of AS and effluent at NLOS2 (14.8% and 18.2%, respectively). Apparently it originates from the influent wastewater supplied for treatment where its share in the resistome was 3.2%. AmpC β-lactamases are considered clinically important cephalosporinases encoded on the chromosomes and plasmids of various bacteria (especially Enterobacteriaceae ), where they mediate resistance to cephalothin, cefazolin, cefoxitin and most penicillins 35 . Close homologues of this gene, with a nucleotide sequence identity of 99.8–100%, have been found in plasmids and chromosomes of various Proteobacteria ( Thauera, Sphingobium, Aeromonas etc.). Since in all samples ampC was found in short contigs with very high coverage, it is likely widespread in the genomes of various bacteria in different genetic contexts.
The second most abundant ARG in the resistomes of AS samples was sulfonamide-resistant dihydropteroate synthase ( sul1 ). It accounted for 4–5% of AS and treated effluent resistomes in LOS and for about 11% in NLOS2, while its share in the influent water resistome was about 5%. The sul1 gene is usually found in class 1 integrons being linked to other resistance genes, including qacE 36 . Consistently, sul1 and qacE were found in one contig assembled for the influent water samples and assigned to Gammaproteobacteria. Another sulfonamide-resistance gene, sul2 , was also numerous, accounting for about 2% of the resistomes in the influent and LOS samples, and for about 4% in the AS and water treated at NLOS2.
Since ARGs entering the activated sludge and then into the treated effluent originate mostly from wastewater supplied for treatment, the absolute majority of ARGs present in the influent in significant amounts (more than 0.2% resistome) in were also found in AS and effluent samples. The only exception macrolide 2′-phosphotransferase gene mph(B) accounting for 0.51% in the influent resistome. Likewise, all ARGs accounting for more than 0.2% of resistomes in the treated effluent were present also in the influent.
Potential multidrug resistant strains
One of the most important public health problems is the spread of multidrug resistant pathogens (MDR), which refers to resistance to at least one agent in three or more chemical classes of antibiotic (e.g. a beta-lactam, an aminoglycoside, a macrolide) 37 . Such strains can arrive with wastewater entering the treatment, and also form in AS communities. AS are dense and highly competitive microbial communities, which, along with the presence of sublethal concentrations of antibiotics and other toxicants in wastewater, creates ideal conditions not only for the selection of resistant strains, but also for the formation of multiple resistance through horizontal gene transfer 4 . To identify MDR bacteria, we binned metagenomic contigs into metagenome-assembled genomes (MAGs) and looked for MAGs comprising several ARGs. Only MAGs with more than 70% completeness and less than 15% contamination were selected for analysis: 117, 56, 72, 94 and 121 for influent, AS of LOS, effluent of LOS, AS of NLOS2 and effluent of NLOS2, respectively. Five MAGs of MDR bacteria were identified in the metagenome of the influent, one—in AS of LOS, two—in the LOS effluent and one in the NLOS2 effluent (Table 3 ). These MAGs were assigned to unclassified genus-level lineages of Ruminococcaceae and Cyclobacteriaceae, Phocaeicola vulgatus, Streptococcus parasuis, Ancrocorticia sp., Enterococcus sp., Bacillus cereus and Undibacterium sp.
Disscussion
We characterized the composition of microbial communities and the resistomes of influent wastewater, activated sludge and treated effluent from two WWTPs in city of Moscow, where various biological water treatment technologies are used. Among the predominant bacteria in the influent wastewater we found mainly fecal contaminants of the genera Collinsella , Bacteroides , Prevotella , Arcobacter , Arcobacteraceae , Blautia , Faecalibacterium, Streptococcus , Acinetobacter , Aeromonas and Veillonella 38 , 39 , 40 , 41 , 42 , 43 . Previously, we performed 16S rRNA gene profiling of wastewater before and after treatment at one WWTP (LOS) and revealed that all abovementioned potential pathogens were efficiently removed and their relative abundance in the water microbiome decreased by 50‒100 times 44 . Similar pattern of removal of potential pathogenic bacteria was observed here for NLOS2 where another water treatment technology is used.
An important indicator of the dissemination of ARG is the proportion of the resistome in the entire metagenome before and after wastewater treatment. In the influent, the resistome accounted for about 0.05% of the metagenome, which corresponds to approximately two ARGs per bacterial genome. Approximately the same values are typical for most countries 3 . After treatment, the fraction of the resistome in the wastewater metagenomes decreases, but, surprisingly, only by 2–4 times. However, since the total concentration of microorganisms in treated effluent is approximately two orders of magnitude lower than in raw wastewater, it is likely that the total abundance of ARGs in the treated effluent is significantly reduced.
Apparently, fecal contaminants effectively removed during treatment are not the only carriers of ARG in wastewater, which are also found in bacteria characteristic of activated sludge and thus appearing in the effluents. Unfortunately, due to the high diversity of microbiomes and the tendency of ARG to be present in multiple copies in different genomic environments, most of the contigs containing ARG turned out to be short, which did not allow to define their taxonomic affiliation.
The resistome of influent water includes 26 ARGs, the share of which is more than 1%. Among of them the prevalence of ampC, aadA, qacE, bla, qacF and qacL is specific for Moscow WWTPs, since these genes were not among the 50 most common ARGs according to the results of a worldwide analysis of wastewater resistomes in large cities 3 . Different ARGs were most “evenly” represented in the influent wastewater while in the AS and treated effluent, a clear selection of particular types of ARGs was observed, which obviously reflects a change in the composition of microbiomes. A vivid example is the increase in the proportion of ampC in the resistomes, especially at LOS.
The discovered ARGs can confer resistance to most classes of antibiotics and among the resistomes of the studied WWTPs in the city of Moscow, genes conferring resistance to beta-lactam antibiotics were the most common, they accounted for about 26% of the resistome in the water supplied for treatment (Fig. 4 ). Similar values have been observed for wastewater in some other countries, particularly in Eastern Europe and Brazil, where 20 to 25% of reads were assigned to ARGs conferring resistance to beta-lactams 3 . According to data for 2021, beta lactams accounted for about 40% of the total antibiotic consumption in Russia in the medical sector 45 .
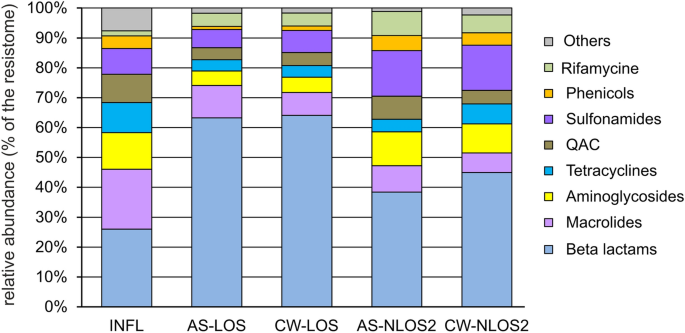
The relative abundancies of ARGs in the resistomes categorized by drug classes.
Like in most wastewater resistomes in different countries, ARGs conferring resistance to macrolides, aminoglycosides and tetracycline were also among the most abundant in wastewater from Moscow (Fig. 4 ). Resistance to macrolides, rather than beta-lactams, was most common in wastewater from most countries in Europe and North America, while in Moscow ARGs to macrolide were the second most common. Macrolides and tetracyclines are also widely used in medicine in Russia (20% and 5% of total antibiotic consumption in 2021, respectively). On the contrary, medical consumption of aminoglycosides in Russia is rather low (< 1% of the total), therefore, the high abundance of relevant ARGs was unexpected. The opposite pattern was observed for quinolones, which make up about 22% of the antibiotics used in medicine, but their ARGs accounted for only about 1% of the resistome. However the main mechanisms of resistance to quinolones, mutations in the target enzymes, DNA gyrase and DNA topoisomerase IV, and increased drug efflux 46 , were not addressed in our study.
A peculiar feature of Moscow wastewater resistome was the high content of resistance genes to sulfonamides (about 9%), which were not among the major genes in wastewater resistomes worldwide 3 . Sulfonamides are synthetic antimicrobial agents that currently have limited use in the human medicine, alone or mainly in combination with trimethoprim (a dihydrofolate reductase inhibitor), in the treatment of uncomplicated respiratory, urinary tract and chlamydia infections 7 , 47 . Different sulfonamide ARGs ( sul1, sul2 and sul3 ) were detected in the wastewater in the some countries, including Denmark, Canada, Spain and China, applying culture dependent, independent and qPCR methods 7 . The opposite picture was observed for streptogramin resistance genes, which were among the ARGs in the majority of resistomes worldwide, but in Moscow wastewater they accounted for less than 1%. This is probably due to the limited use of this drug in Russia.
Another distinguishing feature of the resistome of wastewater in Moscow is the high content of ARGs conferring resistance to quaternary ammonium compounds (QAC), about 9%. It can be explained by the frequent use of these antiseptics in medicine. QACs are active ingredients in more than 200 disinfectants currently recommended for inactivation the SARS-CoV-2 (COVID-19) virus 48 . A recent study showed that the number of QACs used to inactivate the virus in public facilities, hospitals and households increased during the COVID-19 pandemic 49 . Indeed, the results of a study dedicated to the study of wastewater resistome worldwide 3 did not reveal the presence of QAC ARGs in the wastewater, since the samples for this study were collected before the pandemic.
An important issue is the extent to which different water treatment technologies remove ARGs. The effective removal of ARG was primary due to a decrease in the concentration of microorganisms in treated effluent, since the share of resistome in the metagenome after treatment decreased by only 2.6 –3.7 times and the NLOS2 plant appeared to be more effective in this respect. However, compared to LOS, treated effluent at NLOS2 contains approximately twice as much suspended solids, probably due to poorer settling characteristics of the sludge indicated by the higher SVI. Therefore, the overall efficiency of removing ARGs from wastewater at two WWTPs may be similar.
Considering the relative abundances of ARGs in the resistomes, genes conferring resistance to macrolides and tetracyclines were removed more efficiently than beta lactamases, especially ampC , and rifampin ADP-ribosyltransferase genes. The low efficiency of removal of the ampC gene and the increase in its abundance in the resistome after wastewater treatment were previously reported for WWTPs in Germany 50 . Efficient removal of ARGs to macrolides ( ermB, ermF, mph(A), mef(A) ) and tetracyclines ( tet(A), tet(C), tet(Q), tet(W) ) has been reported in a number of studies worldwide 51 . ARGs enabling resistance to sulfonamides, tetracyclines and chloramphenicol were more efficiently removed at LOS than at NLOS2, while the opposite was observed for beta lactamases (Fig. 4 ). The later became the most abundant class of ARGs in the treated effluent.
Metagenomic analysis not only identified resistance genes, but also revealed probable MDR strains based on the analysis of assembled MAGs. We identified 9 such strains in both influent, AS and treated effluent. The real number of MDR strains is probably higher, since only a small fraction of all metagenomic contigs was included in the assembled high quality MAGs.
Phocaeicola vulgatus , (formerly Bacteroides vulgatus ), is a mutualistic anaerobic bacteria commonly found in the human gut microbiome and frequently involved in human infections. The results of whole genome analysis showed presence of blaTEM-1 and blaCMY-2 ARGs, which confers resistant to beta-lactams 52 , 53 . P. vulgatus was also identified as potential host for the transmission of tetracycline ARGs 54 . Streptococcus parasuis is an important zoonotic pathogen that causes primarily meningitis, sepsis, endocarditis, arthritis, and pneumonia in both pigs and humans 55 . A variety of MDR strains of this bacterium have been described. For instance, S. parasuis strain H35 was isolated from a lung sample of a pig in China; several ARGs, including optrA , catQ , erm(B), lsa(E), msr(D), mef(A), mdt(A), tet(M), lnu(B), aadE and two copies of aacA-aphD , were found in the chromosome and cfr(D) was detected on plasmid pH35-cfrD 56 . MDR strain of Bacillus cereus was identified in the effluent water microbiome. This bacterium is known as human pathogen and a common cause of food poisoning with toxin-producing property 57 . Bacillus cereus was isolated from drinking water treatment plant in China and antimicrobial susceptibility testing revealed that it was resistant to cefoxitin, penicillin tetracycline 58 , macrolide-lincosamide-streptogramin (MLSB), aminoglycoside and tetracycline antibiotics 59 . Assembled MAG B.cereus from effluent water contained ARGs conferring to macrolides, beta-lactams, fosfomycin and streptogramin and may be considered as MDR strain. Genomes of members of the genera Streptococcus (AS of LOS) and Enterococcus (influent), not identified at the species level, were found to contain multiple ARGs. Most of species of these genera are opportunistic and true pathogens known for their drug resistance 60 , 61 . One MAG from the influent water metagenome was assigned to uncultured lineage of the family Ruminococcaceae. Members of this family are typical non-pathogenic gut inhabitants, although genomes of some strains could harbor ARGs 62 .
Three MAGs retrieved from influent wastewater microbiome ( Ancrocorticia ) and treated effluent water ( Cyclobacteriaceae and Undibacterium ) were found to contain several ARGs. However, we found no evidences about pathogenic and MDR strains in these taxa. It is possible that these environmental bacteria acquired ARGs via horizontal gene from outside their lineages. WWTPs are an ideal environment for horizontal gene transfer (HGT), since when bacteria are exposed to strong selective pressures, such as the presence of antimicrobials, the horizontal acquisition of ARGs enables genetic diversification and create the potential for rapid gains in fitness 63 .
Conclusions
Metagenome sequencing of the raw wastewater, activated sludge and treated wastewater at two large WWTPs of the Moscow city revealed several hundreds of ARGs that could confer resistance to most commonly used classes of antibiotics.
Resistome accounted for about 0.05% of the wastewater metagenome and after wastewater treatment its share decreased by 3–4 times.
The resistomes were dominated by ARGs encoding resistance to beta-lactams, macrolides, aminoglycosides, tetracycline, QAC, and sulfonamides. A peculiar feature of Moscow wastewater resistome was the high content of ARGs to sulfonamides and limited occurrence of resistance to streptogramins.
ARGs for macrolides and tetracyclines were removed more efficiently than ARGs for beta-lactamases.
A comparison of wastewater resistomes from Moscow and around the world suggested that the abundance and content of ARG in wastewater depend on social, medical, and environmental factors.
Data availability
The raw data generated from 16S rRNA gene sequencing and metagenome sequencing have been deposited in the NCBI Sequence Read Archive (SRA) and are available via the BioProject PRJNA945245.
Cassini, A. et al. Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European Economic Area in 2015: A population-level modelling analysis. Lancet Infect. Dis. 19 , 56–66 (2019).
Article PubMed PubMed Central Google Scholar
Thorpe, K. E., Joski, P. & Johnston, K. J. Antibiotic-resistant infection treatment costs. antibiotic-resistant infection treatment costs have doubled since 2002, now exceeding $2 billion annually. Health Aff. 37 , 662–669 (2018).
Article Google Scholar
Munk, P. et al. Genomic analysis of sewage from 101 countries reveals global landscape of antimicrobial resistance. Nat. Commun. 13 , 7251 (2022).
Article ADS CAS PubMed PubMed Central Google Scholar
Rizzo, L. et al. Urban wastewater treatment plants as hotspots for antibiotic resistant bacteria and genes spread into the environment: a review. Sci. Total Environ. 447 , 345–360 (2013).
Article ADS CAS PubMed Google Scholar
Begmatov, S. et al. The structure of microbial communities of activated sludge of large-scale wastewater treatment plants in the city of Moscow. Sci. Rep. 12 , 3458 (2022).
Mosaka, T. B. M., Unuofin, J. O., Daramola, M. O., Tizaoui, C. & Iwarere, S. A. Inactivation of antibiotic-resistant bacteria and antibiotic-resistance genes in wastewater streams: Current challenges and future perspectives. Front. Microbiol. 13 , 1100102 (2022).
Article PubMed Google Scholar
Pazda, M., Kumirska, J., Stepnowski, P. & Mulkiewicz, E. Antibiotic resistance genes identified in wastewater treatment plant systems—A review. Sci. Total Environ. 697 , 134023 (2019).
Karkman, A., Do, T. T., Walsh, F. & Virta, M. P. J. Antibiotic-resistance genes in waste water. Trends Microbiol. 26 , 220–228 (2018).
Article CAS PubMed Google Scholar
Amos, G. C., Hawkey, P. M., Gaze, W. H. & Wellington, E. M. Waste water effluent contributes to the dissemination of CTX-M-15 in the natural environment. J. Antimicrob Chemother. 69 , 1785–1791 (2014).
Article CAS PubMed PubMed Central Google Scholar
Marti, E., Jofre, J. & Balcazar, J. L. Prevalence of antibiotic resistance genes and bacterial community composition in a river influenced by a wastewater treatment plant. PLoS ONE 8 , e78906 (2013).
Szczepanowski, R. et al. Detection of 140 clinically relevant antibiotic-resistance genes in the plasmid metagenome of wastewater treatment plant bacteria showing reduced susceptibility to selected antibiotics. Microbiology 155 , 2306–2319 (2009).
Mao, D. et al. Prevalence and proliferation of antibiotic resistance genes in two municipal wastewater treatment plants. Water Res. 85 , 458–466 (2015).
Yang, Y., Li, B., Ju, F. & Zhang, T. Exploring variation of antibiotic resistance genes in activated sludge over a four-year period through a metagenomic approach. Environ. Sci. Technol. 47 , 10197–10205 (2013).
Yang, Y., Li, B., Zou, S., Fang, H. H. & Zhang, T. Fate of antibiotic resistance genes in sewage treatment plant revealed by metagenomic approach. Water Res. 62 , 97–106 (2014).
Bengtsson-Palme, J. et al. Elucidating selection processes for antibiotic resistance in sewage treatment plants using metagenomics. Sci. Total Environ. 572 , 697–712 (2016).
Karkman, A. et al. High-throughput quantification of antibiotic resistance genes from an urban wastewater treatment plant. FEMS Microbiol Ecol. 92 , fiw014 (2016).
Laht, M. et al. Abundances of tetracycline, sulphonamide and beta-lactam antibiotic resistance genes in conventional wastewater treatment plants (WWTPs) with different waste load. PLoS ONE 9 , e103705. https://doi.org/10.1371/journal.pone.0103705 (2014).
Auerbach, E. A., Seyfried, E. E. & McMahon, K. D. Tetracycline resistance genes in activated sludge wastewater treatment plants. Water Res. 41 , 1143–1151 (2007).
Harris, S. J., Cormican, M. & Cummins, E. Antimicrobial residues and antimicrobial-resistant bacteria: Impact on the microbial environment and risk to human health—a review. Human Ecol. Risk Assess. Int. J. 18 , 767–809 (2012).
Article CAS Google Scholar
Frey, B. et al. Microbial diversity in European alpine permafrost and active layers. FEMS Microbiol. Ecol. 3 , fiw018 (2016).
Magoc, T. & Salzberg, S. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27 , 2957–2963 (2011).
Edgar, R. C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26 , 2460–2461 (2010).
Rognes, T., Flouri, T., Nichols, B., Quince, C. & Mahé, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ 4 , e2584 (2016).
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17 , 10–12 (2011).
Nurk, S., Meleshko, D., Korobeynikov, A. & Pevzner, P. A. metaSPAdes: A new versatile metagenomic assembler. Genome Res. 27 , 824–834 (2017).
Kang, D. D. et al. MetaBAT 2: an adaptive binning algorithm for robust and efficient genome reconstruction from metagenome assemblies. PeerJ. 7 , e7359 (2019).
Wu, Y. W., Simmons, B. A. & Singer, S. W. MaxBin 2.0: An automated binning algorithm to recover genomes from multiple metagenomic datasets. Bioinformatics. 32 , 605–607 (2016).
Alneberg, J. et al. Binning metagenomic contigs by coverage and composition. Nat. Methods 11 , 1144–1146 (2014).
Sieber, C. M. K. et al. Recovery of genomes from metagenomes via a dereplication, aggregation and scoring strategy. Nat. Microbiol. 3 , 836–843 (2018).
Parks, D. H., Imelfort, M., Skennerton, C. T., Hugenholtz, P. & Tyson, G. W. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2 , 1043–1055 (2015).
Chaumeil, P. A., Hugenholtz, P. & Parks, D. H. GTDB-Tk v2: memory friendly classification with the genome taxonomy database. Bioinformatics 38 , 5315–5316 (2022).
Parks, D. H. et al. A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat. Biotechnol. 36 , 996–1004 (2018).
Hyatt, D. et al. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 11 , 119 (2010).
Feldgarden, M. et al. AMRFinderPlus and the reference gene catalog facilitate examination of the genomic links among antimicrobial resistance, stress response, and virulence. Sci. Rep. 11 , 12728 (2021).
Jacoby, G. A. AmpC beta-lactamases. Clin. Microbiol. Rev. 22 , 161–182 (2009).
Antunes, P., Machado, J., Sousa, J. C. & Peixe, L. Dissemination of sulfonamide resistance genes (sul1, sul2, and sul3) in Portuguese Salmonella enterica strains and relation with integrons. Antimicrob. Agents Chemother. 49 , 836–839 (2005).
Magiorakos, A. P. et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 18 , 268–281 (2012).
Azcarate-Peril, M. A. et al. Impact of short-chain galactooligosaccharides on the gut microbiome of lactose-intolerant individuals. Proc. Natl. Acad. Sci. USA 114 , E367–E375 (2017).
Koskey, A. M. et al. Blautia and Prevotella sequences distinguish human and animal fecal pollution in Brazil surface waters. Environ. Microbiol. Rep. 6 , 696–704 (2014).
Altwegg, M. & Geiss, H. K. Aeromonas as a human pathogen. Crit. Rev. Microbiol. 16 , 253–286 (1989).
Wexler, A. G. & Goodman, A. L. An insider’s perspective: Bacteroides as a window into the microbiome. Nat. Microbiol. 2 , 17026 (2017).
Collado, L. & Figueras, M. J. Taxonomy, epidemiology, and clinical relevance of the genus Arcobacter . Clin. Microbiol. Rev. 24 , 174–192 (2011).
Antunes, L. C., Visca, P. & Towner, K. J. Acinetobacter baumannii : evolution of a global pathogen. Pathog. Dis. 71 , 292–301 (2014).
Begmatov, S. A., Dorofeev, A. G., Pimenov, N. V., Mardanov, A. V. & Ravin, N. V. High efficiency of removal of pathogenic microorganisms at wastewater treatment plants in the city of Moscow. Microbiology 92 , 734–738. https://doi.org/10.1134/S0026261723601124 (2023).
Zakharenkov, I. A., Rachina, S. A., Kozlov, R. S. & Belkova, Yu. A. Consumption of systemic antibiotics in the Russian Federation in 2017–2021. Clin. Microbiol. Antimicrob. Chemother. 24 , 220–225. https://doi.org/10.36488/cmac.2022.3.220-225 (2022).
Hooper, D. C. & Jacoby, G. A. Mechanisms of drug resistance: quinolone resistance. Ann. N.Y. Acad. Sci. 1354 , 12–31 (2015).
Littlefield, B. A., Gurpide, E., Markiewicz, L., McKinley, B. & Hochberg, R. B. A simple and sensitive microtiter plate estrogen bioassay based on stimulation of alkaline phosphatase in Ishikawa cells: estrogenic action of Δ5 adrenal steroids. Endocrinology 127 , 2757–2762 (1990).
Hora, P. I., Pati, S. G., McNamara, P. J. & Arnold, W. A. Increased use of quaternary ammonium compounds during the SARS-CoV-2 pandemic and beyond: consideration of environmental implications. Environ. Sci. Technol. Lett. 7 , 622–631 (2020).
Liu, C. et al. Low concentration quaternary ammonium compounds promoted antibiotic resistance gene transfer via plasmid conjugation. Sci. Total. Environ. 887 , 163781 (2023).
Alexander, J., Bollmann, A., Seitz, W. & Schwartz, T. Microbiological characterization of aquatic microbiomes targeting taxonomical marker genes and antibiotic resistance genes of opportunistic bacteria. Sci. Total. Environ. 512–513 , 316–325 (2015).
Article ADS PubMed Google Scholar
Uluseker, C. et al. A Review on Occurrence and spread of antibiotic resistance in wastewaters and in wastewater treatment plants: mechanisms and perspectives. Front. Microbiol. 12 , 717809 (2021).
Vázquez-López, R. et al. The beta-lactam resistome expressed by aerobic and anaerobic bacteria isolated from human feces of healthy donors. Pharmaceuticals 14 , 533 (2021).
Vishwanath, S., Shenoy, P. A. & Chawla, K. Antimicrobial resistance profile and nim gene detection among Bacteroides fragilis group isolates in a university hospital in South India. J. Glob. Infect. Dis. 11 , 59–62 (2019).
Qin, R., Yu, Q. G., Liu, Z. Y. & Wang, H. Co-occurrence of tetracycline antibiotic resistance genes and microbial communities in plateau wetlands under the influence of human activities. Huan Jing Ke Xue 44 , 169–179 (2023).
PubMed Google Scholar
Wang, J. et al. Investigation of the genomic and pathogenic features of the potentially zoonotic Streptococcus parasuis . Pathogens 10 , 834 (2021).
Zhu, Y. et al. Identification of a Streptococcus parasuis isolate co-harbouring the oxazolidinone resistance genes cfr(D) and optrA . J. Antimicrob. Chemother. 76 , 3059–3061 (2021).
Enosi, T. D., Mathur, A., Ngo, C. & Man, S. M. Bacillus cereus : Epidemiology, virulence factors, and host-pathogen interactions. Trends Microbiol. 29 , 458–471 (2021).
Gu, Q. et al. Characteristics of antibiotic resistance genes and antibiotic-resistant bacteria in full-scale drinking water treatment system using metagenomics and culturing. Front. Microbiol. 12 , 798442 (2022).
Han, Z. et al. Antibiotic resistomes in drinking water sources across a large geographical scale: Multiple drivers and co-occurrence with opportunistic bacterial pathogens. Water Res. 183 , 116088 (2020).
Fiore, E., Van, T. D. & Gilmore, M. S. Pathogenicity of Enterococci . Microbiol. Spectr. https://doi.org/10.1128/microbiolspec.GPP3-0053-2018 (2019).
Patterson, M. J. Streptococcus. In Medical Microbiology 4th edn (ed. Baron, S.) (University of Texas Medical Branch at Galveston, 1996).
Google Scholar
Abdugheni, R. et al. Comparative genomics reveals extensive intra-species genetic divergence of the prevalent gut commensal Ruminococcus gnavus . Microb. Genom. 9 , mgen001071 (2023).
PubMed PubMed Central Google Scholar
Lerminiaux, N. A. & Cameron, A. D. S. Horizontal transfer of antibiotic resistance genes in clinical environments. Can. J. Microbiol. 65 , 34–44 (2019).
Download references
Acknowledgements
This work was partly supported by the Russian Science Foundation (Project 22-74-00022 to S.B.).
Author information
Authors and affiliations.
Institute of Bioengineering, Research Center of Biotechnology of the Russian Academy of Sciences, Leninsky Prosp, bld. 33‑2, Moscow, Russia, 119071
Shahjahon Begmatov, Alexey V. Beletsky, Andrey V. Mardanov & Nikolai V. Ravin
Winogradsky Institute of Microbiology, Research Center of Biotechnology of the Russian Academy of Sciences, Leninsky Prosp, bld. 33‑2, Moscow, Russia, 119071
Alexander G. Dorofeev & Nikolai V. Pimenov
You can also search for this author in PubMed Google Scholar
Contributions
S.B. and N.V.R. designed and supervised the research project; A.G.D. collected the samples and analysed chemical composition of wastewater; A.V.M. performed 16S rRNA gene profiling and metagenome sequencing; S.B., A.V.B., N.V.P., and N.V.R. analysed the sequencing data; S.B. and N.V.R. wrote the manuscript. All authors have read and agreed to the published version of the manuscript.
Corresponding authors
Correspondence to Shahjahon Begmatov or Nikolai V. Ravin .
Ethics declarations
Competing interests.
The authors declare no competing interests.
Additional information
Publisher's note.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Supplementary tables., rights and permissions.
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/ .
Reprints and permissions
About this article
Cite this article.
Begmatov, S., Beletsky, A.V., Dorofeev, A.G. et al. Metagenomic insights into the wastewater resistome before and after purification at large‑scale wastewater treatment plants in the Moscow city. Sci Rep 14 , 6349 (2024). https://doi.org/10.1038/s41598-024-56870-0
Download citation
Received : 26 October 2023
Accepted : 12 March 2024
Published : 15 March 2024
DOI : https://doi.org/10.1038/s41598-024-56870-0
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
By submitting a comment you agree to abide by our Terms and Community Guidelines . If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.
Quick links
- Explore articles by subject
- Guide to authors
- Editorial policies
Sign up for the Nature Briefing: Translational Research newsletter — top stories in biotechnology, drug discovery and pharma.

Study Abroad Aide
The Best Study Abroad Site
Moscow City Teachers’ Training University: Tuition, Admissions, Rankings
Moscow City Teachers’ Training University is a public university in Moscow, Russia. The Ministry of Education established it in 1995 as a pedagogical university, with only 1300 students in its first year. The university currently has over 18,000 students and offers degree programs in the humanities, natural sciences, sports technology, law, business, and language studies.
Moscow City Teachers’ Training University Overview

Moscow City Teachers’ Training University World Rankings
Here are the world rankings of Moscow City Teachers’ Training University from reputable ranking sources:
Ready to take your education to the next level? Discover the top universities in Russia by clicking here .
Tuition Fees at Moscow City Teachers’ Training University
Moscow City Teachers’ Training University tuition fees for both bachelor's and master's students are discussed in this section.
Bachelor's Tuition Fees
Master's tuition fees.
Although this range provide a good estimate of tuition costs at Moscow City Teachers’ Training University, the actual fees depend on your chosen program. Thus, for more exact figures, you may refer to Moscow City Teachers’ Training University tuition fee pages.
Ready to save in tuition and study in Russia? Click here to read our guides to tuition fees and cheapest universities in Russia and start planning your dream education today.
Admissions At Moscow City Teachers’ Training University
If you wish to study at Moscow City Teachers’ Training University, you’ll need to know the admission process, requirements, and other relevant information. Those may vary based on the program you’re applying to Moscow City Teachers’ Training University.
Moreover, the process and requirements for Moscow City Teachers’ Training University international admission may differ from domestic admission. Thus, if you’re an international student, check what additional requirements you need to submit and how you can validate your academic credentials.
To help you find all the admission-related information, we’ve compiled the bachelor's and master's admission pages of Moscow City Teachers’ Training University. Select which one applies to you and take note of the process and requirements.
From choosing your university to applying for your student visa, we know that studying abroad is not easy. However, we are here to support you every step of the way! Don't forget to explore our comprehensive guide as you prepare for your educational journey in Russia.
Degree Programs offered at Moscow City Teachers’ Training University
There are several Moscow City Teachers’ Training University majors or degree programs for bachelor’s and master’s level that you can pursue. Here are some of them:
If you want to know what other academic programs Moscow City Teachers’ Training University offers, please visit the bachelor’s and master’s course pages. These pages will give you detailed information of the programs they offer.
Explore Popular Universities in Russia
Leave a reply cancel reply.
Your email address will not be published. Required fields are marked *
Save my name, email, and website in this browser for the next time I comment.

IMAGES
VIDEO
COMMENTS
RUA: Analyzing Published Research Guidelines Purpose The purpose of this paper is to interpret the two articles identified as most important to the group topic. Course outcomes: This assignment enables the student to meet the following course outcomes. 1. Apply research principles to the interpretation of the content of published research studies.
This assignment enables the student to meet the following course outcomes: CO 2: Apply research principles to the interpretation of the content of published research studies. (POs #4 and #8) CO 4: Evaluate published nursing research for credibility and clinical significance related to evidence- based practice. (POs #4 and #8) DUE DATE
1 Week 5 RUA: Analyzing Published Research Elieana Herrera NR 449: Evidence-Based Practice Chamberlain University September 2021 f2 Clinical Question The RUA assignment delegated to this team is understanding COVID-19 infections in the pediatric populatio. Solutions available. NR 449. Chamberlain College of Nursing.
RUA week 5 Matrix Table Analyzing Published Research; RUA week 3 Topic Search Strategy; Related Studylists Nr 449 NR449 NR449. Preview text. Research on the Role of the Nurse in Using Telemedicine. Your Name Here Chamberlain College of Nursing NR 449: Evidence-Based Practice July 2021.
ANALYZING PUBLISHED RESEARCH 2 PROBLEM The focus of the group's project is to show the correlation between nurse's stress level & the effects on advanced technology on it. The problem described in the research paper by St. Pierre, Alderson& Saint-Jean (2012), To develop & test st&ardized primary health assessment instrument (SPHAI) that can be used to assess the health problems & care ...
Required Uniform Assignment: Analyzing Published Research. Purpose . The purpose of this paper is to interpret the two articles identified as most important to the group topic. ... RUA Analyzing Published Research .docx Revised 07 / 25 /2016 2 . NR449 Evidence Based Practice . NR449 RUA A nalyzing Published Research .docx Revised 07/25 /2016 4 ...
View Homework Help - NR449_RUA_Analyzing_Published_Research.pdf from NR 449 at Chamberlain College of Nursing. NR449 Evidence Based Practice Required Uniform Assignment: Analyzing Published
Analyzing and synthesizing a scholarly journal article is intended to help students obtain the reading and critical thinking skills needed to develop and write their own research papers. This assignment also supports workplace skills where you could be asked to summarize a report or other type of document and report it, for example, during a ...
ANALYZING PUBLISHED RESEARCH Clinical Question The Required Uniform Assignment delegated to our team is Healthcare Associated Infection (HAI). HAI is an infection that a patient received due to procedure and poor interventions. Patients were admitted getting treatment for illnesses and ending up ...
NR 449 Unit 5 Required Uniform Assignment (RUA) 2; Analyzing Published Research. NR 449 Unit 5 Required Uniform Assignment (RUA) 2; Analyzing Published Research. 100% satisfaction guarantee Immediately available after payment Both online and in PDF No strings attached. Previously searched by you.
View NR449_RUA_Analyzing_Published_Research_August_2016.pdf from NR 449 at Chamberlain College of Nursing. NR449 Evidence Based Practice Required Uniform Assignment: Analyzing Published. AI Homework Help. ... 55-60 points Summary omits two or three required items from the Evidence Matrix Table. 51-55 points Summary omits four required items ...
Nr 449 unit 5 required uniform assignment rua 2 analyzing published research. Content type User Generated. Uploaded By ... ANALYZING PUBLISHED RESEARCH Analyzing Published Research Chamberlain College of Nursing Evidence-Based Practice NR449 1 2 ANALYZING PUBLISHED RESEARCH PROBLEM The focus of the group's project is to show the correlation ...
NR 449 Unit 5 Required Uniform Assignment (RUA) 2; Analyzing Published Research. NR 449 Unit 5 Required Uniform Assignment (RUA) 2; Analyzing Published Research. 0. Shopping cart · 0 item · $0.00. Checkout . login ; Sell ; 0. Shopping cart · 0 item · $0.00. Checkout ...
Required Uniform Assignment: Analyzing Published Research . Purpose. ... CO 4: Evaluate published nursing research for credibility and clinical significance related to evidence- based practice. Total Points Possible :200 Points Requirements. The paper will include the following. 1. Clinical question
The fourth considers the augmentation of the methods derived from Design with Nature in reference to innovations in technology since its publication and the contribution that the art of landscape painters can make to landscape analysis and interpretation. The fifth part considers the application of this experience to the international ...
Wastewater treatment plants (WWTPs) are considered to be hotspots for the spread of antibiotic resistance genes (ARGs). We performed a metagenomic analysis of the raw wastewater, activated sludge ...
Required Uniform Assignment: Analyzing Published Research. NR449 Evidence Based Practice. Question: Is there an increased incidence of C-diff in patients of extended stays in the acute care setting compared to patients in a long term care facility? I can provide the full outline for the document with what is being asked and articles
The constructed index is applied to the analysis of Moscow transportation statistics in 2012-2017 provided by the Moscow Traffic Management Center, Yandex and TomTom. Discover the world's ...
The second article by Nymberg, et al., (2019) was more specific to e-health with the elderly than the first, but covered the same issue of how could that population's attitude and confidence with technology affect their health. The Topic Search Strategy paper is the first of three related assignments in this course. It is due in Week 3.
ANALYZING PUBLISHED RESEARCH 2 The focus of the group's project is to show the correlation between nurse's stress level & the effects on advanced technology on it. The problem described in the research paper by St. Pierre, Alderson& Saint-Jean (2012), To develop & test st&ardized primary health assessment instrument (SPHAI) that can be used to assess the health problems & care needs of ...
NR 449 Unit 5 Required Uniform Assignment (RUA) 2; Analyzing Published Research. NR 449 Unit 5 Required Uniform Assignment (RUA) 2; Analyzing Published Research. 100% Money Back Guarantee Immediately available after payment Both online and in PDF No strings attached. Sell. Where do you study.
Student Type. Annual Tuition Fees in RUB. Domestic Students. 107,700 RUB - 165,500 RUB. International Students. 107,700 RUB - 165,500 RUB. Although this range provide a good estimate of tuition costs at Moscow City Teachers' Training University, the actual fees depend on your chosen program. Thus, for more exact figures, you may refer to ...