
International Conference on Intelligent Systems Design and Applications
ISDA 2022: Intelligent Systems Design and Applications pp 374–383 Cite as

A Step-To-Step Guide to Write a Quality Research Article
- Amit Kumar Tyagi ORCID: orcid.org/0000-0003-2657-8700 14 ,
- Rohit Bansal 15 ,
- Anshu 16 &
- Sathian Dananjayan ORCID: orcid.org/0000-0002-6103-7267 17
- Conference paper
- First Online: 01 June 2023
205 Accesses
19 Citations
Part of the book series: Lecture Notes in Networks and Systems ((LNNS,volume 717))
Today publishing articles is a trend around the world almost in each university. Millions of research articles are published in thousands of journals annually throughout many streams/sectors such as medical, engineering, science, etc. But few researchers follow the proper and fundamental criteria to write a quality research article. Many published articles over the web become just irrelevant information with duplicate information, which is a waste of available resources. This is because many authors/researchers do not know/do not follow the correct approach for writing a valid/influential paper. So, keeping such issues for new researchers or exiting researchers in many sectors, we feel motivated to write an article and present some systematic work/approach that can help researchers produce a quality research article. Also, the authors can publish their work in international conferences like CVPR, ICML, NeurIPS, etc., or international journals with high factors or a white paper. Publishing good articles improve the profile of researchers around the world, and further future researchers can refer their work in their work as references to proceed with the respective research to a certain level. Hence, this article will provide sufficient information for researchers to write a simple, effective/impressive and qualitative research article on their area of interest.
- Quality Research
- Research Paper
- Qualitative Research
- Quantitative Research
- Problem Statement
This is a preview of subscription content, log in via an institution .
Buying options
- Available as PDF
- Read on any device
- Instant download
- Own it forever
- Available as EPUB and PDF
- Compact, lightweight edition
- Dispatched in 3 to 5 business days
- Free shipping worldwide - see info
Tax calculation will be finalised at checkout
Purchases are for personal use only
Nair, M.M., Tyagi, A.K., Sreenath, N.: The future with industry 4.0 at the core of society 5.0: open issues, future opportunities and challenges. In: 2021 International Conference on Computer Communication and Informatics (ICCCI), pp. 1–7 (2021). https://doi.org/10.1109/ICCCI50826.2021.9402498
Tyagi, A.K., Fernandez, T.F., Mishra, S., Kumari, S.: Intelligent Automation Systems at the Core of Industry 4.0. In: Abraham, A., Piuri, V., Gandhi, N., Siarry, P., Kaklauskas, A., Madureira, A. (eds.) ISDA 2020. AISC, vol. 1351, pp. 1–18. Springer, Cham (2021). https://doi.org/10.1007/978-3-030-71187-0_1
Chapter Google Scholar
Goyal, D., Tyagi, A.: A Look at Top 35 Problems in the Computer Science Field for the Next Decade. CRC Press, Boca Raton (2020) https://doi.org/10.1201/9781003052098-40
Tyagi, A.K., Meenu, G., Aswathy, S.U., Chetanya, V.: Healthcare Solutions for Smart Era: An Useful Explanation from User’s Perspective. In the Book “Recent Trends in Blockchain for Information Systems Security and Privacy”. CRC Press, Boca Raton (2021)
Google Scholar
Varsha, R., Nair, S.M., Tyagi, A.K., Aswathy, S.U., RadhaKrishnan, R.: The future with advanced analytics: a sequential analysis of the disruptive technology’s scope. In: Abraham, A., Hanne, T., Castillo, O., Gandhi, N., Nogueira Rios, T., Hong, T.-P. (eds.) HIS 2020. AISC, vol. 1375, pp. 565–579. Springer, Cham (2021). https://doi.org/10.1007/978-3-030-73050-5_56
Tyagi, A.K., Nair, M.M., Niladhuri, S., Abraham, A.: Security, privacy research issues in various computing platforms: a survey and the road ahead. J. Inf. Assur. Secur. 15 (1), 1–16 (2020)
Madhav, A.V.S., Tyagi, A.K.: The world with future technologies (Post-COVID-19): open issues, challenges, and the road ahead. In: Tyagi, A.K., Abraham, A., Kaklauskas, A. (eds.) Intelligent Interactive Multimedia Systems for e-Healthcare Applications, pp. 411–452. Springer, Singapore (2022). https://doi.org/10.1007/978-981-16-6542-4_22
Mishra, S., Tyagi, A.K.: The role of machine learning techniques in the Internet of Things-based cloud applications. In: Pal, S., De, D., Buyya, R. (eds.) Artificial Intelligence-Based Internet of Things Systems. Internet of Things (Technology, Communications and Computing). Springer, Cham. https://doi.org/10.1007/978-3-030-87059-1_4
Pramod, A., Naicker, H.S., Tyagi, A.K.: Machine Learning and Deep Learning: Open Issues and Future Research Directions for Next Ten Years. Computational Analysis and Understanding of Deep Learning for Medical Care: Principles, Methods, and Applications. Wiley Scrivener (2020)
Kumari, S., Tyagi, A.K., Aswathy, S.U.: The Future of Edge Computing with Blockchain Technology: Possibility of Threats, Opportunities and Challenges. In the Book Recent Trends in Blockchain for Information Systems Security and Privacy. CRC Press, Boca Raton (2021)
Dananjayan, S., Tang, Y., Zhuang, J., Hou, C., Luo, S.: Assessment of state-of-the-art deep learning based citrus disease detection techniques using annotated optical leaf images. Comput. Electron. Agric. 193 (7), 106658 (2022). https://doi.org/10.1016/j.compag.2021.106658
Nair, M.M., Tyagi, A.K.: Privacy: History, Statistics, Policy, Laws, Preservation and Threat analysis. J. Inf. Assur. Secur. 16 (1), 24–34 (2021)
Tyagi, A.K., Sreenath, N.: A comparative study on privacy preserving techniques for location based services. Br. J. Math. Comput. Sci. 10 (4), 1–25 (2015). ISSN: 2231–0851
Rekha, G., Tyagi, A.K., Krishna Reddy, V.: A wide scale classification of class imbalance problem and its solutions: a systematic literature review. J. Comput. Sci. 15 (7), 886–929 (2019). ISSN Print: 1549–3636
Kanuru, L., Tyagi, A.K., A, S.U., Fernandez, T.F., Sreenath, N., Mishra, S.: Prediction of pesticides and fertilisers using machine learning and Internet of Things. In: 2021 International Conference on Computer Communication and Informatics (ICCCI), pp. 1–6 (2021). https://doi.org/10.1109/ICCCI50826.2021.9402536
Ambildhuke, G.M., Rekha, G., Tyagi, A.K.: Performance analysis of undersampling approaches for solving customer churn prediction. In: Goyal, D., Gupta, A.K., Piuri, V., Ganzha, M., Paprzycki, M. (eds.) Proceedings of the Second International Conference on Information Management and Machine Intelligence. LNNS, vol. 166, pp. 341–347. Springer, Singapore (2021). https://doi.org/10.1007/978-981-15-9689-6_37
Sathian, D.: ABC algorithm-based trustworthy energy-efficient MIMO routing protocol. Int. J. Commun. Syst. 32 , e4166 (2019). https://doi.org/10.1002/dac.4166
Varsha, R., et al.: Deep learning based blockchain solution for preserving privacy in future vehicles. Int. J. Hybrid Intell. Syst. 16 (4), 223–236 (2020)
Tyagi, A.K., Aswathy, S U.: Autonomous Intelligent Vehicles (AIV): research statements, open issues, challenges and road for future. Int. J. Intell. Netw. 2 , 83–102 (2021). ISSN 2666–6030. https://doi.org/10.1016/j.ijin.2021.07.002
Tyagi, A.K., Sreenath, N.: Cyber physical systems: analyses, challenges and possible solutions. Internet Things Cyber-Phys. Syst. 1 , 22–33 (2021). ISSN 2667–3452, https://doi.org/10.1016/j.iotcps.2021.12.002
Tyagi, A.K., Aghila, G.: A wide scale survey on botnet. Int. J. Comput. Appl. 34 (9), 9–22 (2011). (ISSN: 0975–8887)
Tyagi, A.K., Fernandez, T.F., Aswathy, S.U.: Blockchain and aadhaar based electronic voting system. In: 2020 4th International Conference on Electronics, Communication and Aerospace Technology (ICECA), Coimbatore, pp. 498–504 (2020). https://doi.org/10.1109/ICECA49313.2020.9297655
Kumari, S., Muthulakshmi, P.: Transformative effects of big data on advanced data analytics: open issues and critical challenges. J. Comput. Sci. 18 (6), 463–479 (2022). https://doi.org/10.3844/jcssp.2022.463.479
Article Google Scholar
Download references
Acknowledgement
We want to think of the anonymous reviewer and our colleagues who helped us to complete this work.
Author information
Authors and affiliations.
Department of Fashion Technology, National Institute of Fashion Technology, New Delhi, India
Amit Kumar Tyagi
Department of Management Studies, Vaish College of Engineering, Rohtak, India
Rohit Bansal
Faculty of Management and Commerce (FOMC), Baba Mastnath University, Asthal Bohar, Rohtak, India
School of Computer Science and Engineering, Vellore Institute of Technology, Chennai, Tamilnadu, 600127, India
Sathian Dananjayan
You can also search for this author in PubMed Google Scholar
Contributions
Amit Kumar Tyagi & Sathian Dananjayan have drafted and approved this manuscript for final publication.
Corresponding author
Correspondence to Amit Kumar Tyagi .
Editor information
Editors and affiliations.
Faculty of Computing and Data Science, FLAME University, Pune, Maharashtra, India
Ajith Abraham
Center for Smart Computing Continuum, Burgenland, Austria
Sabri Pllana
University of Bari, Bari, Italy
Gabriella Casalino
University of Jinan, Jinan, Shandong, China
Department of Computer Science and Engineering, Thapar Institute of Engineering and Technology, Patiala, Punjab, India
Ethics declarations
Conflict of interest.
The author declares that no conflict exists regarding the publication of this paper.
Scope of the Work
As the author belongs to the computer science stream, so he has tried to cover up this article for all streams, but the maximum example used in situations, languages, datasets etc., are with respect to computer science-related disciplines only. This work can be used as a reference for writing good quality papers for international conferences journals.
Disclaimer. Links and papers provided in the work are only given as examples. To leave any citation or link is not intentional.
Rights and permissions
Reprints and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper.
Tyagi, A.K., Bansal, R., Anshu, Dananjayan, S. (2023). A Step-To-Step Guide to Write a Quality Research Article. In: Abraham, A., Pllana, S., Casalino, G., Ma, K., Bajaj, A. (eds) Intelligent Systems Design and Applications. ISDA 2022. Lecture Notes in Networks and Systems, vol 717. Springer, Cham. https://doi.org/10.1007/978-3-031-35510-3_36
Download citation
DOI : https://doi.org/10.1007/978-3-031-35510-3_36
Published : 01 June 2023
Publisher Name : Springer, Cham
Print ISBN : 978-3-031-35509-7
Online ISBN : 978-3-031-35510-3
eBook Packages : Intelligent Technologies and Robotics Intelligent Technologies and Robotics (R0)
Share this paper
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Publish with us
Policies and ethics
- Find a journal
- Track your research
How to write a research article to submit for publication
Pharmacists and healthcare professionals who are undertaking research should have an understanding of how to produce a research article for publication, and be aware of the important considerations relating to submission to a peer-reviewed journal.

Viennaslide / Alamy Stock Photo
Undertaking and performing scientific, clinical and practice-based research is only the beginning of the scholarship of discovery [1] . For the full impact of any research to be achieved and to have an effect on the wider research and scientific community, it must be published in an outlet accessible to relevant professionals [2] .
Scientific research is often published in peer-reviewed journals. Peer review is defined as the unbiased, independent, critical assessment of scholarly or research manuscripts submitted to journals by experts or opinion leaders [3] . The process and requirements of reviewers has been covered recently [4] . On account of this rigorous process, peer-reviewed scientific journals are considered the primary source of new information that impacts and advances clinical decision-making and practice [5] , [6] .
The development of a research article can be helpful for the promotion of scientific thinking [7] , [8] and the advancement of effective writing skills, allowing the authors to participate in broader scientific discussions that lie beyond their scope of practice or discipline [2] .
This article aims to provide pharmacists and healthcare professionals who are undertaking research with an understanding of how to produce a research article for publication, as well as points to consider before submission to a peer-reviewed journal.
Importance of the research question
This article will not go into detail about forming suitable research questions, however, in principle, a good research question will be specific, novel and of relevance to the scientific community (e.g. pharmacy – pharmacists, pharmaceutical scientists, pharmacy technicians and related healthcare professionals). Hulley et al . suggest using the FINER criteria (see ‘Figure 1: FINER criteria for a good research question’) to aid the development of a good research question [9] .
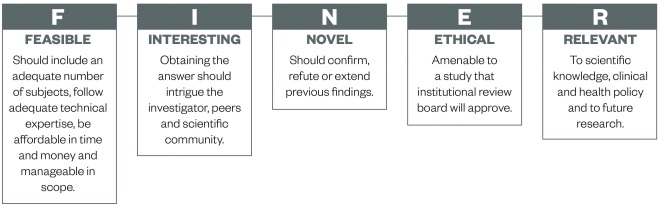
Figure 1: FINER criteria for a good research question
Source: Hulley S, Cummings S, Browner W et al . [9]
The FINER criteria highlight useful points that may generally increase the chances of developing a successful research project. A good research question should specify the population of interest, be of interest to the scientific community and potentially to the public, have clinical relevance and further current knowledge in the field.
Having a clear research question that is of interest to those working in the same field will help in the preparation of an article because it can be used as the central organising principle – all of the content included and discussed should focus on answering this question.
Preparing a first draft
Before writing the article, it is useful to highlight several journals that you could submit the final article to. It also helps to familiarise yourself with these journals’ styles, article structures and formatting instructions before starting to write. Many journals also have criteria that research articles should be able to satisfy. For example, all research article submissions to Clinical Pharmacist must demonstrate innovative or novel results that are sustainable, reproducible and transferable [10] .
Having researched potential target journals, you should have a clear idea about your target audience, enabling you to pitch the level of the article appropriately [11] (see ‘Box 1: Top tips to prepare for writing’).
Box 1: Top tips to prepare for writing
- Know the focus of the paper – identify two or three important findings and make these the central theme of the article;
- Gather important data, perform any analyses and make rough data plots and tables beforehand. These can then be refined for inclusion or submitted as supplementary information if needed;
- Organise your results to flow in a logical sequence;
- Know the structure and requirements of your target journals (check websites and author guidelines, as well as published articles);
- Think about the style of the piece and look to pitch the article at the level of the intended audience;
- Clarity should be your guiding principle.
Structuring a research article
Most research articles follow a similar structure and format that includes an abstract, introduction, methods, results and discussion, as well as a summary of the key points discussed in the article.
One approach is to start with the methods section, which can be derived from the protocol and any pilot phase. Many of the figures and tables can be constructed in advance, which will help with writing the results section. The questions addressed by the study can be used alongside the results to formulate the introduction, which can guide how the discussion is written [11] .
Clinical Pharmacist , like other peer-reviewed journals, has specific author guidelines and formatting instructions to help authors prepare their articles [10] , [12] , [13] . The following sections will discuss the required sections and important considerations for authors when writing.
Title, abstract and keywords
The title, abstract and keywords are essential to the successful communication of research. Most electronic search engines, databases (e.g. PubMed/MEDLINE) and journal websites extract words from them to determine whether your article will be displayed to interested readers [14] , [15] , [16] , [17] , enabling accurate dissemination and leading to future citations.
In addition, the title and abstract are usually freely available online. If an article is not published in an ‘open access’ format, (i.e. it is free and immediately available online and access is combined with the rights to use these articles fully in the digital environment) [18] , or if the reader does not have a subscription to the journal, they will have to decide on whether to pay to access the full article to continue reading. Therefore, it is imperative that they are informative and accurate.
The title should accurately reflect the research, identify the main issue and begin with the subject matter, while being both simple and enticing enough to attract the audience [19] . Authors should avoid using ‘a study of’, ‘investigations into’ and ‘observations on’ in titles. It is also worth remembering that abstracting and indexing services, such as MEDLINE, require accurate titles, because they extract keywords from them for cross-referencing [19] .
Many journals require the abstract to be structured in the same way as the main headings of the paper (e.g. introduction, methods, results, discussion and conclusion) and to be around 150–300 words in length [10] . In general, references should not be cited in the abstract.
Introduction
The introduction should provide the background and context to the study. Two or three paragraphs can be dedicated to the discussion of any previous work and identification of gaps in current knowledge. The rest of the introduction should then outline what this piece of work aims to address and why this is important, before stating the objectives of the study and the research question [20] .
The methods section should provide the reader with enough detail for them to be able to reproduce the study if desired [3] . The context and setting of the study should be described and the study design specified. The section should further describe the population (including the inclusion and exclusion criteria), sampling strategy and the interventions performed. The main study variables should be identified and the data collection procedures described [3] .
Authors should provide specific, technical and detailed information in this section. Several checklists and guidelines are available for the reporting of specific types of studies:
- CONSORT is used for developing and reporting a randomised controlled trial [21] ;
- The STARD checklist can help with designing a diagnostic accuracy study [22] ;
- The PRISMA checklist can be used when performing a metaâ€analyses or systematic review, but can also help with compiling an introduction [23] .
For the reporting of qualitative research studies, authors should explain which research tradition the study utilises and link the choice of methodological strategies with the research goals [24] .
For studies describing the development of new initiatives or clinical services, authors should describe the situation before the initiative began, the establishment of priorities, formulation of objectives and strategies, mobilisation of resources, and processes used in the methods section [10] .
The final portion of the methods section will include the statistical methods used to analyse the data [25] . The statistical methods employed should be described with enough detail to enable a knowledgeable reader with access to the original data to be able to judge its appropriateness for the study and verify the results [3] . For survey-based studies and information on sampling frame, size and statistical powers, see ‘When to use a survey in pharmacy practice research’ [26] .
Findings should be quantified and presented with appropriate indicators of measurement error or uncertainty (e.g. confidence intervals). Authors should avoid relying solely on statistical hypothesis testing, such as P values, because these fail to convey important information about effect size and precision of estimates [3] . Statistical terms, abbreviations and most symbols should be defined, and the statistical software package and versions used should be specified. Authors should also take care to distinguish prespecified from exploratory analyses, including subgroup analyses [3] .
The results section should be straightforward and factual and all of the results that relate to the research question should be provided, with detail including simple counts and percentages [27] . Data collection and recruitment should be commented on and the participants described. Secondary findings and the results of subgroup analyses can also be presented [27] .
Figures, schemes and tables
To present data and results of the research study, figures, schemes and tables can be used. They should include significant digits, error bars and levels of statistical significance.
Tables should be presented with a summary title, followed by caption, a sentence or two that describes the content and impact of the data included in the table. All captions should provide enough detail so that the table or figure can be interpreted and understood as stand-alone material, separate from the article.
Figures should also be presented with a summary title, a caption that describes the significant result or interpretation that can be made from the figure, the number of repetitions within the experiment, as well as what the data point actually represents. All figures and tables should be cited in the manuscript text [11] .
When compiling tables and figures, important statistics, such as the number of samples (n), the index of dispersion (standard deviation [SD], standard error of the mean [SEM]), and the index of central tendency (mean, median or mode), must be stated. The statistical analysis performed should also be included and specific statistical data should be indicated (e.g. P values) [11] .
Discussion and conclusions
The discussion section should state the main findings of the study. The main results should be compared with reference to previous research and current knowledge, and where this has been extended it should be fully described [2] , [11] , [25] . For clinical studies, relevant discussion of the implications the results may have on policy should be included [10] . It is important to include an analysis of the strengths and limitations of the study and offer perspectives for future work [2] . Excessive presentation of data and results without any discussion should be avoided and it is not necessary to cite a published work for each argument presented. Any conclusions should include the major findings, followed by a brief discussion of future perspectives and the application of this work to other disciplines [10] .
The list of references should be appropriate; important statements presented as facts should be referenced, as well as the methods and instruments used. Reference lists for research articles, however, unlike comprehensive reviews of a topic, do not necessarily have to be exhaustive. References to unpublished work, to documents in the grey literature (technical reports), or to any source that the reader will have difficulty finding or understanding should be avoided [27] . Most journals have reference limits and specific formatting requirements, so it is important to check the journal’s author guidelines [10] , [11] , [12] , [13] , [19] .
Authorship and acknowledgements
Determining contributors who qualify as authors and those who should be acknowledged can be difficult. Clinical Pharmacist follows guidance from the International Committee of Medical Journal Editors, which recommends that authorship be based on the following four criteria:
- Substantial contributions to the conception or design of the work; or the acquisition, analysis, or interpretation of data for the work; AND
- Drafting the work or revising it critically for important intellectual content; AND
- Final approval of the version to be published; AND
- Agreement to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved [3] .
Therefore, only individuals who meet all four criteria should be identified as authors [3] . The contribution of individuals who do not meet all four criteria should instead be included in the acknowledgements.
In addition, a statement that recognises any funding sources for the work should be added to the acknowledgements. This statement should adhere to the guidelines provided by the funding institution [11] .
Supplementary and supporting information
A key principle of research publication is that others should be able to replicate and build upon an author’s published claims. Therefore, submitted manuscripts should contain the necessary detail about the study and analytical design, and the data must be available for editors and peer-reviewers to allow full evaluation to take place. This is now commonplace and is seen as best practice. Author guidelines now include sections related to misconduct and falsification of data [28] . By participating in self-archiving practices and providing full data sets, authors can play their part in transparency.
The Royal Pharmaceutical Society website hosts a database to help share data from research studies. The map of evidence collates existing evidence and ongoing initiatives that can ultimately inform policy and practice relating to pharmacy; enables the sharing and showcasing of good pharmacy practice and innovation; and aims to increase the knowledge exchange and learning in pharmacy and pharmaceutical sciences [29] .
Revising your article prior to submission
Once a draft research article has been prepared, it should be shared among all of the co-authors for review and comments. A full revision of the draft should then take place to correct grammar and check flow and logic before journal submission. All authors will have to agree on the authenticity of the data and presentation before formal submission can take place [3] (see ‘Box 2: Common mistakes and reasons why research articles are rejected for publication’).
Box 2: Common mistakes and reasons why research articles are rejected for publication
- Lack of novelty and importance of the research question;
- Poor study design;
- Methods not accurately and adequately described;
- Results poorly reported, along with little analysis of data;
- Lack of statistical analysis;
- Not acknowledging the study’s limitations;
- Providing unsupported conclusions or overstating the results of the study;
- Poor writing;
- Not following the journal’s style and formatting guidance;
- Submitting a manuscript that is incomplete or outside of the aims and scope.
Selecting a journal and submitting your manuscript
It is important to select a journal for submission wisely because this choice can determine the impact and dissemination of your work [13] . Impact factor (a measure of the frequency with which the average article in a journal has been cited in a particular year), the scope and readership of a title may also influence your choice.
Furthermore, approval and adequate disclosures must be obtained from all co-authors. A conflict of interests form is also completed as part of the submissions process (normally completed by the lead author on behalf of all authors).
Many journals now request that a cover letter is also submitted to the editor, putting the study in context and explaining why the research is of importance to their audience and why it should be considered for publication in their journal.
Once this is all completed, the article can be formally submitted (usually via email or an online submission system). Figure 2 provides a sample process for a manuscript once submitted to a journal for consideration for publication.
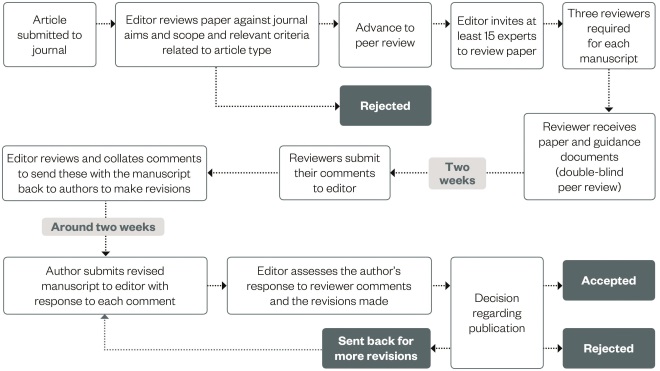
Figure 2: Sample process for a submitted manuscript
Source: The Pharmaceutical Journal
All journals follow a similar process for article submissions, whether they use a formal online submissions system or simply email. Clinical Pharmacist uses a process similar to this and it is useful for authors to be aware of how their submission may progress once submitted to a journal for publication.
Useful Links
- The EQUATOR Network
- Research4Life – Authorship skills modules
- Pharmacy Research UK
Reading this article counts towards your CPD
You can use the following forms to record your learning and action points from this article from Pharmaceutical Journal Publications.
Your CPD module results are stored against your account here at The Pharmaceutical Journal . You must be registered and logged into the site to do this. To review your module results, go to the ‘My Account’ tab and then ‘My CPD’.
Any training, learning or development activities that you undertake for CPD can also be recorded as evidence as part of your RPS Faculty practice-based portfolio when preparing for Faculty membership. To start your RPS Faculty journey today, access the portfolio and tools at www.rpharms.com/Faculty
If your learning was planned in advance, please click:
If your learning was spontaneous, please click:
[1] Boyer E. Scholarship reconsidered: Priorities for the professoriate . 1990. Princeton, NJ: The Carnegie Foundation for the Advancement of Teaching.
[2] Hoogenboom BJ & Manske RC. How to write a scientific article. Int J Sports Phys Ther . 2012;7(5):512–517. PMCID: PMC3474301
[3] International Committee of Medical Journal Editors. Recommendations for the conduct, reporting, editing, and publication of scholarly work in medical journals. 2014. Available at: http://www.icmje.org/icmje-recommendations.pdf (accessed November 2016).
[4] Dowdall M. How to be an effective peer reviewer. Clinical Pharmacist 2015;7(10). doi: 10.1211/CP.2015.20200006
[5] Nahata MC. Tips for writing and publishing an article. Ann Pharmaco . 2008;42:273–277. doi: 10.1345/aph.1K616
[6] Dixon N. Writing for publication: A guide for new authors. Int J Qual Health Care . 2001;13:417–421. doi: 10.1093/intqhc/13.5.417
[7] Keys CW. Revitalizing instruction in scientific genres: Connecting knowledge production with writing to learn in science. Sci Educ . 1999;83:115–130.
[8] Gopen G & Swan J. The science of scientific writing. Am Sci . 1990;78:550–558. Available at: http://www.americanscientist.org/issues/pub/the-science-of-scientific-writing (accessed November 2016)
[9] Hulley S, Cummings S, Browner W et al . Designing clinical research. 3rd ed. Philadelphia (PA): Lippincott Williams and Wilkins; 2007.
[10] The Pharmaceutical Journal and Clinical Pharmacist. Author Guidance (2015). Available at: http://www.pharmaceutical-journal.com/for-authors-and-referees/article-types/#Practice_reports (accessed November 2016)
[11] Fisher JP, Jansen JA, Johnson PC et al . Guidelines for writing a research article for publication. Mary Ann Liebert Inc. Available at: https://www.liebertpub.com/media/pdf/English-Research-Article-Writing-Guide.pdf (accessed November 2016)
[12] Nature. Author Resources: How to write a paper. Available at: http://www.nature.com/authors/author_resources/how_write.html (accessed November 2016)
[13] Wiley Online Library. Resources for authors and reviewers: preparing your article. Available at: http://olabout.wiley.com/WileyCDA/Section/id-828006.html (accessed November 2016)
[14] SAGE Publications. Help readers find your article. Available at: http://www.uk.sagepub.com/journalgateway/findArticle.htm (accessed November 2016)
[15] Bem DJ. Writing the empirical journal article. In: MP Zanna & JM Darley (Eds.), The complete academic: a practical guide for the beginning social scientist (pp. 171–201). New York: Random House; 1987.
[16] Fathalla M & Fathalla M. A practical guide for health researchers. Available at: http://www.emro.who.int/dsaf/dsa237.pdf (accessed November 2016)
[17] Coghill A & Garson L (Eds.). Scientific Papers. In: A Coghill & L Garson (Eds.), The ACS Style Guide, 3 rd Edition (pp. 20–21). New York: Oxford University Press, 2006.
[18] The Scholarly Publishing and Academic Resources Institute. Available at: http://sparcopen.org/open-access/ (accessed November 2016).
[19] Elsevier. Understanding the publishing process: how to publish in scholarly journals. Available at: https://www.elsevier.com/__data/assets/pdf_file/0003/91173/Brochure_UPP_April2015.pdf (accessed November 2016).
[20] SciDevNet. How do I write a scientific paper? 2008. Available at: http://www.scidev.net/global/publishing/practical-guide/how-do-i-write-a-scientific-paper-.html (accessed November 2016)
[21] Moher D, Schultz KR & Altman DG. CONSORT GROUP (Consolidatied Standards of Reporting Trials). The CONSORT statement: Revised recommendations for improving the quality of reports of parallelâ€group randomized controlled trials. Ann Intern Med . 2001;134:657–662. PMID: 11304106
[22] Bossuyt PM, Reitsma JB, Bruns DE et al . Towards complete and accurate reporting of studies of diagnostic accuracy: The STARD Initiative. Ann Int Med 2003;138:40–44. PMID: 12513043
[23] Moher D, Liberati A, Tetzlaff J et al . The PRISMA Group (2009). Preferred reporting items for systematic reviews and metaâ€analyses: the PRISMA statement. PLoS Med 6(6):e1000097. doi: 10.1371/journal.pmed1000097
[24] Devers KJ & Frankel RM. Getting qualitative research published. Educ Health 2001;14:109–117. doi: 10.1080/13576280010021888
[25] Van Way CW. Writing a scientific paper. Nutr Clin Pract 2007;22:636–640. PMID: 18042951
[26] Kishore V. When to use a survey in pharmacy practice research. The Pharmaceutical Journal 296(7886). doi: 10.1211/PJ.2016.20200700
[27] Perneger PV & Hudelson PM. Writing a research article: advice to beginners . Int J Qual Health Care 2004;16(3):191–192. doi: 10.1093/intqhc/mzh053
[28] World Association of Medical Editors. Professionalism Code of Conduct. 2016. Available at: http://www.wame.org/News/Details/16 (accessed November 2016)
[29] Royal Pharmaceutical Society. Map of Evidence. Available at: http://www.rpharms.com/support/map-of-evidence.asp (accessed November 2016)
You might also be interested in…

Working to improve our digital archive
The Pharmaceutical Journal is moving forward into a digital future

The launch of our new digital platform is just the beginning of our plans for the future of PJ
Typical structure of a research paper
This Feature Is Available To Subscribers Only
Sign In or Create an Account
This PDF is available to Subscribers Only
For full access to this pdf, sign in to an existing account, or purchase an annual subscription.
An official website of the United States government
The .gov means it’s official. Federal government websites often end in .gov or .mil. Before sharing sensitive information, make sure you’re on a federal government site.
The site is secure. The https:// ensures that you are connecting to the official website and that any information you provide is encrypted and transmitted securely.
- Publications
- Account settings
Preview improvements coming to the PMC website in October 2024. Learn More or Try it out now .
- Advanced Search
- Journal List
- J Hum Reprod Sci
- v.10(1); Jan-Mar 2017
Preparing and Publishing a Scientific Manuscript
Padma r. jirge.
Department of Reproductive Medicine, Sushrut Assisted Conception Clinic Shreyas Hospital, Kolhapur, Maharashtra, India
Publishing original research in a peer-reviewed and indexed journal is an important milestone for a scientist or a clinician. It is an important parameter to assess academic achievements. However, technical and language barriers may prevent many enthusiasts from ever publishing. This review highlights the important preparatory steps for creating a good manuscript and the most widely used IMRaD (Introduction, Materials and Methods, Results, and Discussion) method for writing a good manuscript. It also provides a brief overview of the submission and review process of a manuscript for publishing in a biomedical journal.
B ACKGROUND
T he publication of original research in a peer-reviewed and indexed journal is the ultimate and most important step toward the recognition of any scientific work. However, the process starts long before the write-up of a manuscript. The journal in which the author wishes to publish his/her work should be chosen at the time of conceptualization of the scientific work based on the expected readership.
The journals do provide information on the “scope of the journal,” which specifies the scientific areas relevant for publication in the journal, and “instructions to authors,” which need to be adhered to while preparing a manuscript.
The publication of scientific work has become mandatory for scientists or specialists holding academic affiliations, and it is now desirable even at an undergraduate level. Despite a plethora of forums for presenting the original research work, very little of it ever gets published in a scientific journal, and even if it does, the manuscripts are usually from the same few institutions.[ 1 , 2 ] It serves the purpose of academic recognition; and certain publications may even contribute to shaping various national policies. An academic appointment, suitable infrastructure, and access to peer-reviewed journals are considered as the facilitators for publishing.[ 3 ]
The lack of technical and writing skills, institutional hurdles, and time constraints are considered as the major hurdles for any scientific publication.[ 3 ] In addition, the majority of clinicians in India are involved in providing healthcare in the private sector in individually owned hospitals or those governed by small groups of doctors. This necessitates performing a multitude of tasks apart from providing core clinical care and, hence, poses an additional limiting factor because of the long and irregular working hours.
It is extremely challenging to dedicate some time for research and writing in such a scenario. However, it is a loss to science if this group of skilled clinicians does not contribute to medical literature.
Maintaining the ethics and science of research and understanding the norms of preparing a manuscript are very important in improving the quality and relevance of clinical research in our country. This article brings together various aspects to be borne in mind while creating a manuscript suitable for publication. The inputs provided are relevant to all those interested, irrespective of whether they have an academic or institutional affiliation. While the prospect of becoming an author of a published scientific work is exciting, it is important to be prepared for minor or major revisions in the original article and even rejection. However, persevering in this endeavor may help preserving one’s work and contribute to the promotion of science.[ 4 , 5 ]
Important considerations for writing a manuscript include the following:
- (1) Conceptualization of a clinically relevant scientific work.
- (2) Choosing an appropriate journal and an alternative one.
- (3) Familiarizing with instructions to authors.
- (4) Coordination and well-defined task delegation within the team and involvement of a biostatistician from the conception of the study.
- (5) Preparing a skeletal framework for writing the manuscript.
- (6) Delegating time for thinking and writing at regular intervals.
S TEPS I NVOLVED IN M ANUSCRIPT P REPARATION
A manuscript should both be informative and readable. Even though the concept is clear in the authors’ mind, it is important to remember that they are introducing some new work for the readers, and, hence, appropriate organization of the manuscript is necessary to make the purpose and importance of the work clear to the readers.
- (1) Choosing the appropriate journal for publication : The preferred choice of journal should be one of the first steps to be considered, as mentioned earlier. The guidelines for authors may change with time and, hence, should be referred to at regular intervals and conformed to. The choice of journal principally depends on the target readers, and it may be necessary to have one or more journals in mind in case of nonacceptance from the journal of first choice. A journal’s impact factor is to be considered while choosing an appropriate journal.
Majority of the biomedical journals with good impact factor have specific authorship criteria.[ 8 ] This prevents problems related to ghost authorship and honorary authorship. Ghost authorship refers to a scenario wherein an author’s name is omitted to hide financial relationships with private companies; honorary authorship is naming someone who has not made substantial contribution to the work, either due to pressure from colleagues or to improve the chances of publication.[ 9 ]
Most of the journals conform to the authorship criteria defined by the International Committee of Medical Journal Editors.[ 10 ] They are listed as the following:
Substantial contributions to the conception or design of the work; or the acquisition, analysis, or interpretation of data for the work; ANDDrafting the work or revising it critically for important intellectual content; ANDFinal approval of the version to be published; ANDAgreement to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Some journals require authors to declare their contributions to the research work and manuscript preparation. This helps to prevent honorary and ghost authorship and encourages authors to be more honest and accountable.[ 11 ]
Keywords : are mentioned at the bottom of the Abstract section. These words denote the important aspects of the manuscript and help identify the manuscripts by electronic search engines. Most of the journals specify the number of keywords required, usually between 4 and 8. They need to be simple and specific to the manuscript; a good title contains majority of the keywords.
The general flow of the manuscript follows an IMRaD (Introduction, Materials and Methods, Results, and Discussion) structure. Even though this has been recommended since the early 20 th century, most of the authors started following it since the 1970s.[ 13 ]
Important components of the Introduction section

A common error while writing an introduction is an attempt to review the entire evidence available on the topic. This becomes confusing to the reader, and the purpose and importance of the study in question gets submerged in the plethora of information provided. Issues mentioned in the Introduction section will need to be addressed in the Discussion section, and it is important to avoid repetitions and overlapping. Some may prefer to write the Introduction section after preparing the draft of the Materials and Methods and Results sections.
The last paragraph in the Introduction section defines the aim of the study or the study question using active verbs. If there is more than one aim for the study, specify the primary aim and address the secondary aims in a separate sentence. It is recommended that the Introduction section should not occupy more than 10–15% of the entire text.[ 14 ]
This is followed by a detailed description of the study protocol. At times, some of the methods used may be very elaborate and not very relevant to majority of the readers, for example, if polymerase chain reaction (PCR) is used for diagnosis, the type of PCR performed should be mentioned in this section, but the entire procedure need not be elaborated in the “methods” section. Either a relevant reference can be provided or the procedural details can be given online as supplemental data.
It is important to mention both the generic and brand names of all the drugs used along with the name of the manufacturer and the place of manufacturing. Similarly, all the hematological, biochemical, hormonal assays, and radiological investigations performed should provide the specifications of the equipment used and its manufacturer’s details. For many biochemical and endocrine parameters, it is preferred that the intra- and interassay coefficients of variation are provided. In addition, the standard units of measurements and the internationally accepted abbreviations should be used.[ 18 ]
There are online guidelines available to maintain uniformity in reporting the different types of studies such as Consolidated Standards of Reporting Trials (CONSORT) for randomized controlled trials, Strengthening the Reporting of Observational studies in Epidemiology (STROBE) for observational studies, and Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) for systematic reviews.[ 19 ] Adherence to these guidelines improves the clarity and completeness of reporting.
Statistical analysis : One of the most important deterrents for publishing clinical research is the inability to choose and perform appropriate statistical analysis. With the availability of various user-friendly software systems, an increasing number of the researchers are comfortable performing complex analyses without additional assistance. However, it is still a common practice to involve biostatisticians for this purpose. Coordination between the clinicians and biostatisticians is very important for sample size calculation, creation of a proper data set, and its subsequent analysis. It is important to use the appropriate statistical methodologies for a more complete representation of the data to improve the quality of a manuscript.[ 20 ] It may be helpful to refer to a recent review of the most widely used statistical analyses and their application in clinical research for a better data presentation.[ 20 ] There is some evidence that structured training involving data analysis, manuscript writing, and submission to indexed journals improves the quality of submitted manuscripts even in a low-resource setting.[ 21 ] Short, online certificate courses on biostatistics are available free of cost from many universities across the globe. The important aspects regarding the Materials and Methods section are summarized in Table 2 .
Important components of the Materials and Methods section

The results of the study are summarized in the form of tables and figures. Journals may have limitations on the number of figures and tables, as well as the rows and columns in tables. The text should only highlight the findings recorded in the tables and figures and should not repeat every detail.[ 16 ] Primary analysis should be presented in a separate paragraph. Any secondary analysis performed in view of the results seen in the primary analysis should be mentioned separately [ Table 3 ].
Important components of the Results section

When comparing two groups, it is a good practice to mention the data pertaining to the study group followed by that of the control group and to maintain the same order throughout the section. No adjectives should be used while comparing, except for the statistical significance of the findings. The Results section is written in the past tense, and the numerical values should be presented with a maximum of one decimal place.
Statistical significance as shown by P-value, if accompanied by odds ratio and 95% confidence interval gives important information of direction and size of treatment effect. The measures of central tendencies should be followed by the appropriate measures of variability (mean and standard deviation; median and interquartile range). Relative measures should be accompanied by absolute values (percentage and actual value).[ 22 ] The interpretation of results solely based on bar diagrams or line graphs could be misleading, and a more complete data may be presented in the form of box plots or scatter plots.[ 20 ]
The strengths and weaknesses of the study should be discussed in a separate paragraph. This makes way for implications for clinical practice and future research.[ 16 , 23 ]
The section ends with a conclusion of not more than one to two sentences. The Conclusion section summarizes the study findings in the context of evidence in the field. The important components of the Discussion section are summarized in Table 4 [ Figure 1 ].
Important components of the Discussion section

The hourglass structure of the Introduction and Discussion sections
A referencing tool such as EndNote™ may be used to store and organize the references. The references at the end of the manuscript need to be listed in a manner specified by the journal. The common styles used are Vancouver, Harvard, APA, etc.[ 24 ] Despite continued efforts, standardization to one global format has not yet become a reality.[ 25 ]
It is important to understand the evidence in the referenced articles to write meaningful Introduction and Discussion sections. Online search engines such as Pubmed, Medline, and Scopus are some of the sources that provide abstracts from indexed journals. However, a full-text article may not always be available unless one has subscription for the journals. Those with institutional attachments, authors, and even the research division of pharmaceutical companies may be unconventional but helpful sources for procuring full-text articles. Individual articles can be purchased from certain journals as well.
- (9) Acknowledgements : This section follows the Conclusion section. People who have helped in various aspects of the concerned research work, statistical analysis, or manuscript preparation, but do not qualify to be authors for the study, are acknowledged, preferably with their academic affiliations.[ 26 ]
The aforementioned section provides the general guidelines for preparing a good manuscript. However, an exhaustive list of available guidelines and other resources to facilitate good research reporting are provided by the Enhancing the Quality and Transparency of Health Research network ( http://www.equator-network.org ).
A DDITIONAL F ACTORS I NFLUENCING THE M ANUSCRIPT Q UALITY
- (1) Plagiarism : Plagiarism is a serious threat to scientific publications and is described by the office of Research Integrity as “theft or misappropriation of intellectual property and the substantial unattributed textual copying of another’s work and the representation of them as one’s own original work.” The primary responsibility of preventing plagiarism lies with the authors. It is important to develop the skill of writing any manuscript in one’s own words and when quoting available evidence, substantiate with appropriate references. However, the use of plagiarism detection tools and a critical analysis by the editorial team prior to submitting an article for peer review are also equally important to prevent this menace.[ 29 ] The consequences of plagiarism could range from disciplinary charges such as retraction of the article to criminal charges.[ 30 ]
- (2) Language : One of the important limitations to publication is the problem of writing in English. This can be minimized by seeking help from colleagues or using the language editing service provided by many of the journals.
- (3) Professional medical writing support : In recent years, it is acknowledged that the lack of time and linguistic constraints prevent some of the good work from being published. Hence, the role of professional medical writing support is being critically evaluated. Declared professional medical writing support is found to be associated with more complete reporting of clinical trial results and higher quality of written English. Medical writing support may play an important role in raising the quality of clinical trial reporting.[ 31 ] The role of professional medical writers should be acknowledged in the Acknowledgements section.[ 32 ]
S UBMISSION TO J OURNALS AND R EVIEWING P ROCESS
The submission of manuscripts is now exclusively an online exercise. The basic model of submission in any journal comprises the following: the title file or first page file, article file, image files, videos, charts, tables, figures, and copyright/consent forms. It is important to keep all the files ready in a folder before starting the submission process. When submitting images, it is important to have good quality, well-focused images with good resolution.[ 33 ] Some journals may offer the choice of selecting preferred reviewers to the authors and hence, one must be prepared for this. Once the manuscript is submitted, the status can be periodically checked. With minor variations, a submitted article goes through the following review process: The Editor allocates it to one of the editorial team members who checks for the suitability for publication in the journal. It is checked for plagiarism as well at this stage. The article then goes for peer review to two to three reviewers. The review process may take 4–6 weeks, at the end of which, the reviewers submit their remarks, and “article decision” is made, which could be an advice for minor/major revisions, rewriting the whole manuscript for specific reasons, acceptance without any changes (very rare), or rejection. It is important to take into consideration all the comments of the reviewers and incorporate the necessary changes in the manuscript before resubmitting. However, if the manuscript is rejected, revise to incorporate the valid suggestions given by the reviewers and consider submitting to another journal in the field. This should be effected without delay overcoming the disappointment so that the research still remains valid in the context of time.
P REDATORY J OURNALS
Some of the well-known journals provide an “open access” option to the authors, wherein if the manuscript is published, it is accessible to all the readers online free of cost. However, the authors need to pay a certain fee to make their manuscript an open access article. In addition, some of the well-known journals published by reputed publishers such as BioMed Central (BMC) and Public Library of Science (PLoS) have online “open access” journals, where the manuscripts are published for a fee but are subjected to the conventional scrutiny process, and the readers can access the full-text article.[ 34 ] The Directory of Open Access Journals, http://doaj.org , is an online directory that indexes and provides access to high-quality, open access, peer-reviewed journals. However, many online open access journals are mushrooming, which provide a legitimate face for an illegitimate publication process lacking basic industry standards, sound peer review practices, and solid basis in publication ethics. Such journals are known as “predatory journals.”[ 35 ] The pressure of needing to have scientific publications and the lack of knowledge regarding predatory journals may encourage authors to submit their articles to such journals. Currently, it is not easy to identify predatory journals, and authors should seek such information proactively from mentors, journal websites, and recent and relevant published literature. In addition, editorial oversights (editors and editorial board members), peer review practices, the quality of published articles, indexing, access, citations and ethical practices are important aspects to be considered while choosing an appropriate journal.[ 36 ]
A relevant research hypothesis and research conducted within the ethical framework are of utmost importance for clinical research. The natural progression from here is the manuscript preparation, a daunting process for most of the clinicians involved in clinical research. Choosing a journal that provides an appropriate platform for the manuscript, conforming to the instructions specific for the journal, and following certain simple guidelines can result in successful preparation and publishing of scientific work. Allocating certain time at regular intervals for writing and maintaining discipline and perseverance in this regard are very important prerequisites to achieve the goal of successful publication.
Financial support and sponsorship
Conflicts of interest.
There are no conflicts of interest.
R EFERENCES
Log in using your username and password
- Search More Search for this keyword Advanced search
- Latest content
- Current issue
- Browse by collection
- BMJ Journals More You are viewing from: Google Indexer
You are here
- Online First
- The role of COVID-19 vaccines in preventing post-COVID-19 thromboembolic and cardiovascular complications
- Article Text
- Article info
- Citation Tools
- Rapid Responses
- Article metrics
- Núria Mercadé-Besora 1 , 2 , 3 ,
- Xintong Li 1 ,
- Raivo Kolde 4 ,
- Nhung TH Trinh 5 ,
- Maria T Sanchez-Santos 1 ,
- Wai Yi Man 1 ,
- Elena Roel 3 ,
- Carlen Reyes 3 ,
- http://orcid.org/0000-0003-0388-3403 Antonella Delmestri 1 ,
- Hedvig M E Nordeng 6 , 7 ,
- http://orcid.org/0000-0002-4036-3856 Anneli Uusküla 8 ,
- http://orcid.org/0000-0002-8274-0357 Talita Duarte-Salles 3 , 9 ,
- Clara Prats 2 ,
- http://orcid.org/0000-0002-3950-6346 Daniel Prieto-Alhambra 1 , 9 ,
- http://orcid.org/0000-0002-0000-0110 Annika M Jödicke 1 ,
- Martí Català 1
- 1 Pharmaco- and Device Epidemiology Group, Health Data Sciences, Botnar Research Centre, NDORMS , University of Oxford , Oxford , UK
- 2 Department of Physics , Universitat Politècnica de Catalunya , Barcelona , Spain
- 3 Fundació Institut Universitari per a la recerca a l'Atenció Primària de Salut Jordi Gol i Gurina (IDIAPJGol) , IDIAP Jordi Gol , Barcelona , Catalunya , Spain
- 4 Institute of Computer Science , University of Tartu , Tartu , Estonia
- 5 Pharmacoepidemiology and Drug Safety Research Group, Department of Pharmacy, Faculty of Mathematics and Natural Sciences , University of Oslo , Oslo , Norway
- 6 School of Pharmacy , University of Oslo , Oslo , Norway
- 7 Division of Mental Health , Norwegian Institute of Public Health , Oslo , Norway
- 8 Department of Family Medicine and Public Health , University of Tartu , Tartu , Estonia
- 9 Department of Medical Informatics, Erasmus University Medical Center , Erasmus University Rotterdam , Rotterdam , Zuid-Holland , Netherlands
- Correspondence to Prof Daniel Prieto-Alhambra, Pharmaco- and Device Epidemiology Group, Health Data Sciences, Botnar Research Centre, NDORMS, University of Oxford, Oxford, UK; daniel.prietoalhambra{at}ndorms.ox.ac.uk
Objective To study the association between COVID-19 vaccination and the risk of post-COVID-19 cardiac and thromboembolic complications.
Methods We conducted a staggered cohort study based on national vaccination campaigns using electronic health records from the UK, Spain and Estonia. Vaccine rollout was grouped into four stages with predefined enrolment periods. Each stage included all individuals eligible for vaccination, with no previous SARS-CoV-2 infection or COVID-19 vaccine at the start date. Vaccination status was used as a time-varying exposure. Outcomes included heart failure (HF), venous thromboembolism (VTE) and arterial thrombosis/thromboembolism (ATE) recorded in four time windows after SARS-CoV-2 infection: 0–30, 31–90, 91–180 and 181–365 days. Propensity score overlap weighting and empirical calibration were used to minimise observed and unobserved confounding, respectively.
Fine-Gray models estimated subdistribution hazard ratios (sHR). Random effect meta-analyses were conducted across staggered cohorts and databases.
Results The study included 10.17 million vaccinated and 10.39 million unvaccinated people. Vaccination was associated with reduced risks of acute (30-day) and post-acute COVID-19 VTE, ATE and HF: for example, meta-analytic sHR of 0.22 (95% CI 0.17 to 0.29), 0.53 (0.44 to 0.63) and 0.45 (0.38 to 0.53), respectively, for 0–30 days after SARS-CoV-2 infection, while in the 91–180 days sHR were 0.53 (0.40 to 0.70), 0.72 (0.58 to 0.88) and 0.61 (0.51 to 0.73), respectively.
Conclusions COVID-19 vaccination reduced the risk of post-COVID-19 cardiac and thromboembolic outcomes. These effects were more pronounced for acute COVID-19 outcomes, consistent with known reductions in disease severity following breakthrough versus unvaccinated SARS-CoV-2 infection.
- Epidemiology
- PUBLIC HEALTH
- Electronic Health Records
Data availability statement
Data may be obtained from a third party and are not publicly available. CPRD: CPRD data were obtained under the CPRD multi-study license held by the University of Oxford after Research Data Governance (RDG) approval. Direct data sharing is not allowed. SIDIAP: In accordance with current European and national law, the data used in this study is only available for the researchers participating in this study. Thus, we are not allowed to distribute or make publicly available the data to other parties. However, researchers from public institutions can request data from SIDIAP if they comply with certain requirements. Further information is available online ( https://www.sidiap.org/index.php/menu-solicitudesen/application-proccedure ) or by contacting SIDIAP ([email protected]). CORIVA: CORIVA data were obtained under the approval of Research Ethics Committee of the University of Tartu and the patient level data sharing is not allowed. All analyses in this study were conducted in a federated manner, where analytical code and aggregated (anonymised) results were shared, but no patient-level data was transferred across the collaborating institutions.
This is an open access article distributed in accordance with the Creative Commons Attribution 4.0 Unported (CC BY 4.0) license, which permits others to copy, redistribute, remix, transform and build upon this work for any purpose, provided the original work is properly cited, a link to the licence is given, and indication of whether changes were made. See: https://creativecommons.org/licenses/by/4.0/ .
https://doi.org/10.1136/heartjnl-2023-323483
Statistics from Altmetric.com
Request permissions.
If you wish to reuse any or all of this article please use the link below which will take you to the Copyright Clearance Center’s RightsLink service. You will be able to get a quick price and instant permission to reuse the content in many different ways.
WHAT IS ALREADY KNOWN ON THIS TOPIC
COVID-19 vaccines proved to be highly effective in reducing the severity of acute SARS-CoV-2 infection.
While COVID-19 vaccines were associated with increased risk for cardiac and thromboembolic events, such as myocarditis and thrombosis, the risk of complications was substantially higher due to SARS-CoV-2 infection.
WHAT THIS STUDY ADDS
COVID-19 vaccination reduced the risk of heart failure, venous thromboembolism and arterial thrombosis/thromboembolism in the acute (30 days) and post-acute (31 to 365 days) phase following SARS-CoV-2 infection. This effect was stronger in the acute phase.
The overall additive effect of vaccination on the risk of post-vaccine and/or post-COVID thromboembolic and cardiac events needs further research.
HOW THIS STUDY MIGHT AFFECT RESEARCH, PRACTICE OR POLICY
COVID-19 vaccines proved to be highly effective in reducing the risk of post-COVID cardiovascular and thromboembolic complications.
Introduction
COVID-19 vaccines were approved under emergency authorisation in December 2020 and showed high effectiveness against SARS-CoV-2 infection, COVID-19-related hospitalisation and death. 1 2 However, concerns were raised after spontaneous reports of unusual thromboembolic events following adenovirus-based COVID-19 vaccines, an association that was further assessed in observational studies. 3 4 More recently, mRNA-based vaccines were found to be associated with a risk of rare myocarditis events. 5 6
On the other hand, SARS-CoV-2 infection can trigger cardiac and thromboembolic complications. 7 8 Previous studies showed that, while slowly decreasing over time, the risk for serious complications remain high for up to a year after infection. 9 10 Although acute and post-acute cardiac and thromboembolic complications following COVID-19 are rare, they present a substantial burden to the affected patients, and the absolute number of cases globally could become substantial.
Recent studies suggest that COVID-19 vaccination could protect against cardiac and thromboembolic complications attributable to COVID-19. 11 12 However, most studies did not include long-term complications and were conducted among specific populations.
Evidence is still scarce as to whether the combined effects of COVID-19 vaccines protecting against SARS-CoV-2 infection and reducing post-COVID-19 cardiac and thromboembolic outcomes, outweigh any risks of these complications potentially associated with vaccination.
We therefore used large, representative data sources from three European countries to assess the overall effect of COVID-19 vaccines on the risk of acute and post-acute COVID-19 complications including venous thromboembolism (VTE), arterial thrombosis/thromboembolism (ATE) and other cardiac events. Additionally, we studied the comparative effects of ChAdOx1 versus BNT162b2 on the risk of these same outcomes.
Data sources
We used four routinely collected population-based healthcare datasets from three European countries: the UK, Spain and Estonia.
For the UK, we used data from two primary care databases—namely, Clinical Practice Research Datalink, CPRD Aurum 13 and CPRD Gold. 14 CPRD Aurum currently covers 13 million people from predominantly English practices, while CPRD Gold comprises 3.1 million active participants mostly from GP practices in Wales and Scotland. Spanish data were provided by the Information System for the Development of Research in Primary Care (SIDIAP), 15 which encompasses primary care records from 6 million active patients (around 75% of the population in the region of Catalonia) linked to hospital admissions data (Conjunt Mínim Bàsic de Dades d’Alta Hospitalària). Finally, the CORIVA dataset based on national health claims data from Estonia was used. It contains all COVID-19 cases from the first year of the pandemic and ~440 000 randomly selected controls. CORIVA was linked to the death registry and all COVID-19 testing from the national health information system.
Databases included sociodemographic information, diagnoses, measurements, prescriptions and secondary care referrals and were linked to vaccine registries, including records of all administered vaccines from all healthcare settings. Data availability for CPRD Gold ended in December 2021, CPRD Aurum in January 2022, SIDIAP in June 2022 and CORIVA in December 2022.
All databases were mapped to the Observational Medical Outcomes Partnership Common Data Model (OMOP CDM) 16 to facilitate federated analytics.
Multinational network staggered cohort study: study design and participants
The study design has been published in detail elsewhere. 17 Briefly, we used a staggered cohort design considering vaccination as a time-varying exposure. Four staggered cohorts were designed with each cohort representing a country-specific vaccination rollout phase (eg, dates when people became eligible for vaccination, and eligibility criteria).
The source population comprised all adults registered in the respective database for at least 180 days at the start of the study (4 January 2021 for CPRD Gold and Aurum, 20 February 2021 for SIDIAP and 28 January 2021 for CORIVA). Subsequently, each staggered cohort corresponded to an enrolment period: all people eligible for vaccination during this time were included in the cohort and people with a history of SARS-CoV-2 infection or COVID-19 vaccination before the start of the enrolment period were excluded. Across countries, cohort 1 comprised older age groups, whereas cohort 2 comprised individuals at risk for severe COVID-19. Cohort 3 included people aged ≥40 and cohort 4 enrolled people aged ≥18.
In each cohort, people receiving a first vaccine dose during the enrolment period were allocated to the vaccinated group, with their index date being the date of vaccination. Individuals who did not receive a vaccine dose comprised the unvaccinated group and their index date was assigned within the enrolment period, based on the distribution of index dates in the vaccinated group. People with COVID-19 before the index date were excluded.
Follow-up started from the index date until the earliest of end of available data, death, change in exposure status (first vaccine dose for those unvaccinated) or outcome of interest.
COVID-19 vaccination
All vaccines approved within the study period from January 2021 to July 2021—namely, ChAdOx1 (Oxford/AstraZeneca), BNT162b2 (BioNTech/Pfizer]) Ad26.COV2.S (Janssen) and mRNA-1273 (Moderna), were included for this study.
Post-COVID-19 outcomes of interest
Outcomes of interest were defined as SARS-CoV-2 infection followed by a predefined thromboembolic or cardiac event of interest within a year after infection, and with no record of the same clinical event in the 6 months before COVID-19. Outcome date was set as the corresponding SARS-CoV-2 infection date.
COVID-19 was identified from either a positive SARS-CoV-2 test (polymerase chain reaction (PCR) or antigen), or a clinical COVID-19 diagnosis, with no record of COVID-19 in the previous 6 weeks. This wash-out period was imposed to exclude re-recordings of the same COVID-19 episode.
Post-COVID-19 outcome events were selected based on previous studies. 11–13 Events comprised ischaemic stroke (IS), haemorrhagic stroke (HS), transient ischaemic attack (TIA), ventricular arrhythmia/cardiac arrest (VACA), myocarditis/pericarditis (MP), myocardial infarction (MI), heart failure (HF), pulmonary embolism (PE) and deep vein thrombosis (DVT). We used two composite outcomes: (1) VTE, as an aggregate of PE and DVT and (2) ATE, as a composite of IS, TIA and MI. To avoid re-recording of the same complication we imposed a wash-out period of 90 days between records. Phenotypes for these complications were based on previously published studies. 3 4 8 18
All outcomes were ascertained in four different time periods following SARS-CoV-2 infection: the first period described the acute infection phase—that is, 0–30 days after COVID-19, whereas the later periods - which are 31–90 days, 91–180 days and 181–365 days, illustrate the post-acute phase ( figure 1 ).
- Download figure
- Open in new tab
- Download powerpoint
Study outcome design. Study outcomes of interest are defined as a COVID-19 infection followed by one of the complications in the figure, within a year after infection. Outcomes were ascertained in four different time windows after SARS-CoV-2 infection: 0–30 days (namely the acute phase), 31–90 days, 91–180 days and 181–365 days (these last three comprise the post-acute phase).
Negative control outcomes
Negative control outcomes (NCOs) were used to detect residual confounding. NCOs are outcomes which are not believed to be causally associated with the exposure, but share the same bias structure with the exposure and outcome of interest. Therefore, no significant association between exposure and NCO is to be expected. Our study used 43 different NCOs from previous work assessing vaccine effectiveness. 19
Statistical analysis
Federated network analyses.
A template for an analytical script was developed and subsequently tailored to include the country-specific aspects (eg, dates, priority groups) for the vaccination rollout. Analyses were conducted locally for each database. Only aggregated data were shared and person counts <5 were clouded.
Propensity score weighting
Large-scale propensity scores (PS) were calculated to estimate the likelihood of a person receiving the vaccine based on their demographic and health-related characteristics (eg, conditions, medications) prior to the index date. PS were then used to minimise observed confounding by creating a weighted population (overlap weighting 20 ), in which individuals contributed with a different weight based on their PS and vaccination status.
Prespecified key variables included in the PS comprised age, sex, location, index date, prior observation time in the database, number of previous outpatient visits and previous SARS-CoV-2 PCR/antigen tests. Regional vaccination, testing and COVID-19 incidence rates were also forced into the PS equation for the UK databases 21 and SIDIAP. 22 In addition, least absolute shrinkage and selection operator (LASSO) regression, a technique for variable selection, was used to identify additional variables from all recorded conditions and prescriptions within 0–30 days, 31–180 days and 181-any time (conditions only) before the index date that had a prevalence of >0.5% in the study population.
PS were then separately estimated for each staggered cohort and analysis. We considered covariate balance to be achieved if absolute standardised mean differences (ASMDs) were ≤0.1 after weighting. Baseline characteristics such as demographics and comorbidities were reported.
Effect estimation
To account for the competing risk of death associated with COVID-19, Fine-and-Grey models 23 were used to calculate subdistribution hazard ratios (sHRs). Subsequently, sHRs and confidence intervals were empirically calibrated from NCO estimates 24 to account for unmeasured confounding. To calibrate the estimates, the empirical null distribution was derived from NCO estimates and was used to compute calibrated confidence intervals. For each outcome, sHRs from the four staggered cohorts were pooled using random-effect meta-analysis, both separately for each database and across all four databases.
Sensitivity analysis
Sensitivity analyses comprised 1) censoring follow-up for vaccinated people at the time when they received their second vaccine dose and 2) considering only the first post-COVID-19 outcome within the year after infection ( online supplemental figure S1 ). In addition, comparative effectiveness analyses were conducted for BNT162b2 versus ChAdOx1.

Supplemental material
Data and code availability.
All analytic code for the study is available in GitHub ( https://github.com/oxford-pharmacoepi/vaccineEffectOnPostCovidCardiacThromboembolicEvents ), including code lists for vaccines, COVID-19 tests and diagnoses, cardiac and thromboembolic events, NCO and health conditions to prioritise patients for vaccination in each country. We used R version 4.2.3 and statistical packages survival (3.5–3), Empirical Calibration (3.1.1), glmnet (4.1-7), and Hmisc (5.0–1).
Patient and public involvement
Owing to the nature of the study and the limitations regarding data privacy, the study design, analysis, interpretation of data and revision of the manuscript did not involve any patients or members of the public.
All aggregated results are available in a web application ( https://dpa-pde-oxford.shinyapps.io/PostCovidComplications/ ).
We included over 10.17 million vaccinated individuals (1 618 395 from CPRD Gold; 5 729 800 from CPRD Aurum; 2 744 821 from SIDIAP and 77 603 from CORIVA) and 10.39 million unvaccinated individuals (1 640 371; 5 860 564; 2 588 518 and 302 267, respectively). Online supplemental figures S2-5 illustrate study inclusion for each database.
Adequate covariate balance was achieved after PS weighting in most studies: CORIVA (all cohorts) and SIDIAP (cohorts 1 and 4) did not contribute to ChAdOx1 subanalyses owing to sample size and covariate imbalance. ASMD results are accessible in the web application.
NCO analyses suggested residual bias after PS weighting, with a majority of NCOs associated positively with vaccination. Therefore, calibrated estimates are reported in this manuscript. Uncalibrated effect estimates and NCO analyses are available in the web interface.
Population characteristics
Table 1 presents baseline characteristics for the weighted populations in CPRD Aurum, for illustrative purposes. Online supplemental tables S1-25 summarise baseline characteristics for weighted and unweighted populations for each database and comparison. Across databases and cohorts, populations followed similar patterns: cohort 1 represented an older subpopulation (around 80 years old) with a high proportion of women (57%). Median age was lowest in cohort 4 ranging between 30 and 40 years.
- View inline
Characteristics of weighted populations in CPRD Aurum database, stratified by staggered cohort and exposure status. Exposure is any COVID-19 vaccine
COVID-19 vaccination and post-COVID-19 complications
Table 2 shows the incidence of post-COVID-19 VTE, ATE and HF, the three most common post-COVID-19 conditions among the studied outcomes. Outcome counts are presented separately for 0–30, 31–90, 91–180 and 181–365 days after SARS-CoV-2 infection. Online supplemental tables S26-36 include all studied complications, also for the sensitivity and subanalyses. Similar pattern for incidences were observed across all databases: higher outcome rates in the older populations (cohort 1) and decreasing frequency with increasing time after infection in all cohorts.
Number of records (and risk per 10 000 individuals) for acute and post-acute COVID-19 cardiac and thromboembolic complications, across cohorts and databases for any COVID-19 vaccination
Forest plots for the effect of COVID-19 vaccines on post-COVID-19 cardiac and thromboembolic complications; meta-analysis across cohorts and databases. Dashed line represents a level of heterogeneity I 2 >0.4. ATE, arterial thrombosis/thromboembolism; CD+HS, cardiac diseases and haemorrhagic stroke; VTE, venous thromboembolism.
Results from calibrated estimates pooled in meta-analysis across cohorts and databases are shown in figure 2 .
Reduced risk associated with vaccination is observed for acute and post-acute VTE, DVT, and PE: acute meta-analytic sHR are 0.22 (95% CI, 0.17–0.29); 0.36 (0.28–0.45); and 0.19 (0.15–0.25), respectively. For VTE in the post-acute phase, sHR estimates are 0.43 (0.34–0.53), 0.53 (0.40–0.70) and 0.50 (0.36–0.70) for 31–90, 91–180, and 181–365 days post COVID-19, respectively. Reduced risk of VTE outcomes was observed in vaccinated across databases and cohorts, see online supplemental figures S14–22 .
Similarly, the risk of ATE, IS and MI in the acute phase after infection was reduced for the vaccinated group, sHR of 0.53 (0.44–0.63), 0.55 (0.43–0.70) and 0.49 (0.38–0.62), respectively. Reduced risk associated with vaccination persisted for post-acute ATE, with sHR of 0.74 (0.60–0.92), 0.72 (0.58–0.88) and 0.62 (0.48–0.80) for 31–90, 91–180 and 181–365 days post-COVID-19, respectively. Risk of post-acute MI remained lower for vaccinated in the 31–90 and 91–180 days after COVID-19, with sHR of 0.64 (0.46–0.87) and 0.64 (0.45–0.90), respectively. Vaccination effect on post-COVID-19 TIA was seen only in the 181–365 days, with sHR of 0.51 (0.31–0.82). Online supplemental figures S23-31 show database-specific and cohort-specific estimates for ATE-related complications.
Risk of post-COVID-19 cardiac complications was reduced in vaccinated individuals. Meta-analytic estimates in the acute phase showed sHR of 0.45 (0.38–0.53) for HF, 0.41 (0.26–0.66) for MP and 0.41 (0.27–0.63) for VACA. Reduced risk persisted for post-acute COVID-19 HF: sHR 0.61 (0.51–0.73) for 31–90 days, 0.61 (0.51–0.73) for 91–180 days and 0.52 (0.43–0.63) for 181–365 days. For post-acute MP, risk was only lowered in the first post-acute window (31–90 days), with sHR of 0.43 (0.21–0.85). Vaccination showed no association with post-COVID-19 HS. Database-specific and cohort-specific results for these cardiac diseases are shown in online supplemental figures S32-40 .
Stratified analyses by vaccine showed similar associations, except for ChAdOx1 which was not associated with reduced VTE and ATE risk in the last post-acute window. Sensitivity analyses were consistent with main results ( online supplemental figures S6-13 ).
Figure 3 shows the results of comparative effects of BNT162b2 versus ChAdOx1, based on UK data. Meta-analytic estimates favoured BNT162b2 (sHR of 0.66 (0.46–0.93)) for VTE in the 0–30 days after infection, but no differences were seen for post-acute VTE or for any of the other outcomes. Results from sensitivity analyses, database-specific and cohort-specific estimates were in line with the main findings ( online supplemental figures S41-51 ).
Forest plots for comparative vaccine effect (BNT162b2 vs ChAdOx1); meta-analysis across cohorts and databases. ATE, arterial thrombosis/thromboembolism; CD+HS, cardiac diseases and haemorrhagic stroke; VTE, venous thromboembolism.
Key findings
Our analyses showed a substantial reduction of risk (45–81%) for thromboembolic and cardiac events in the acute phase of COVID-19 associated with vaccination. This finding was consistent across four databases and three different European countries. Risks for post-acute COVID-19 VTE, ATE and HF were reduced to a lesser extent (24–58%), whereas a reduced risk for post-COVID-19 MP and VACA in vaccinated people was seen only in the acute phase.
Results in context
The relationship between SARS-CoV-2 infection, COVID-19 vaccines and thromboembolic and/or cardiac complications is tangled. Some large studies report an increased risk of VTE and ATE following both ChAdOx1 and BNT162b2 vaccination, 7 whereas other studies have not identified such a risk. 25 Elevated risk of VTE has also been reported among patients with COVID-19 and its occurrence can lead to poor prognosis and mortality. 26 27 Similarly, several observational studies have found an association between COVID-19 mRNA vaccination and a short-term increased risk of myocarditis, particularly among younger male individuals. 5 6 For instance, a self-controlled case series study conducted in England revealed about 30% increased risk of hospital admission due to myocarditis within 28 days following both ChAdOx1 and BNT162b2 vaccines. However, this same study also found a ninefold higher risk for myocarditis following a positive SARS-CoV-2 test, clearly offsetting the observed post-vaccine risk.
COVID-19 vaccines have demonstrated high efficacy and effectiveness in preventing infection and reducing the severity of acute-phase infection. However, with the emergence of newer variants of the virus, such as omicron, and the waning protective effect of the vaccine over time, there is a growing interest in understanding whether the vaccine can also reduce the risk of complications after breakthrough infections. Recent studies suggested that COVID-19 vaccination could potentially protect against acute post-COVID-19 cardiac and thromboembolic events. 11 12 A large prospective cohort study 11 reports risk of VTE after SARS-CoV-2 infection to be substantially reduced in fully vaccinated ambulatory patients. Likewise, Al-Aly et al 12 suggest a reduced risk for post-acute COVID-19 conditions in breakthrough infection versus SARS-CoV-2 infection without prior vaccination. However, the populations were limited to SARS-CoV-2 infected individuals and estimates did not include the effect of the vaccine to prevent COVID-19 in the first place. Other studies on post-acute COVID-19 conditions and symptoms have been conducted, 28 29 but there has been limited reporting on the condition-specific risks associated with COVID-19, even though the prognosis for different complications can vary significantly.
In line with previous studies, our findings suggest a potential benefit of vaccination in reducing the risk of post-COVID-19 thromboembolic and cardiac complications. We included broader populations, estimated the risk in both acute and post-acute infection phases and replicated these using four large independent observational databases. By pooling results across different settings, we provided the most up-to-date and robust evidence on this topic.
Strengths and limitations
The study has several strengths. Our multinational study covering different healthcare systems and settings showed consistent results across all databases, which highlights the robustness and replicability of our findings. All databases had complete recordings of vaccination status (date and vaccine) and are representative of the respective general population. Algorithms to identify study outcomes were used in previous published network studies, including regulatory-funded research. 3 4 8 18 Other strengths are the staggered cohort design which minimises confounding by indication and immortal time bias. PS overlap weighting and NCO empirical calibration have been shown to adequately minimise bias in vaccine effectiveness studies. 19 Furthermore, our estimates include the vaccine effectiveness against COVID-19, which is crucial in the pathway to experience post-COVID-19 complications.
Our study has some limitations. The use of real-world data comes with inherent limitations including data quality concerns and risk of confounding. To deal with these limitations, we employed state-of-the-art methods, including large-scale propensity score weighting and calibration of effect estimates using NCO. 19 24 A recent study 30 has demonstrated that methodologically sound observational studies based on routinely collected data can produce results similar to those of clinical trials. We acknowledge that results from NCO were positively associated with vaccination, and estimates might still be influenced by residual bias despite using calibration. Another limitation is potential under-reporting of post-COVID-19 complications: some asymptomatic and mild COVID-19 infections might have not been recorded. Additionally, post-COVID-19 outcomes of interest might be under-recorded in primary care databases (CPRD Aurum and Gold) without hospital linkage, which represent a large proportion of the data in the study. However, results in SIDIAP and CORIVA, which include secondary care data, were similar. Also, our study included a small number of young men and male teenagers, who were the main population concerned with increased risks of myocarditis/pericarditis following vaccination.
Conclusions
Vaccination against SARS-CoV-2 substantially reduced the risk of acute post-COVID-19 thromboembolic and cardiac complications, probably through a reduction in the risk of SARS-CoV-2 infection and the severity of COVID-19 disease due to vaccine-induced immunity. Reduced risk in vaccinated people lasted for up to 1 year for post-COVID-19 VTE, ATE and HF, but not clearly for other complications. Findings from this study highlight yet another benefit of COVID-19 vaccination. However, further research is needed on the possible waning of the risk reduction over time and on the impact of booster vaccination.
Ethics statements
Patient consent for publication.
Not applicable.
Ethics approval
The study was approved by the CPRD’s Research Data Governance Process, Protocol No 21_000557 and the Clinical Research Ethics committee of Fundació Institut Universitari per a la recerca a l’Atenció Primària de Salut Jordi Gol i Gurina (IDIAPJGol) (approval number 4R22/133) and the Research Ethics Committee of the University of Tartu (approval No. 330/T-10).
Acknowledgments
This study is based in part on data from the Clinical Practice Research Datalink (CPRD) obtained under licence from the UK Medicines and Healthcare products Regulatory Agency. We thank the patients who provided these data, and the NHS who collected the data as part of their care and support. All interpretations, conclusions and views expressed in this publication are those of the authors alone and not necessarily those of CPRD. We would also like to thank the healthcare professionals in the Catalan healthcare system involved in the management of COVID-19 during these challenging times, from primary care to intensive care units; the Institut de Català de la Salut and the Program d’Analítica de Dades per a la Recerca i la Innovació en Salut for providing access to the different data sources accessible through The System for the Development of Research in Primary Care (SIDIAP).
- Pritchard E ,
- Matthews PC ,
- Stoesser N , et al
- Lauring AS ,
- Tenforde MW ,
- Chappell JD , et al
- Pistillo A , et al
- Duarte-Salles T , et al
- Hansen JV ,
- Fosbøl E , et al
- Chen A , et al
- Hippisley-Cox J ,
- Mei XW , et al
- Duarte-Salles T ,
- Fernandez-Bertolin S , et al
- Ip S , et al
- Bowe B , et al
- Prats-Uribe A ,
- Feng Q , et al
- Campbell J , et al
- Herrett E ,
- Gallagher AM ,
- Bhaskaran K , et al
- Raventós B ,
- Fernández-Bertolín S ,
- Aragón M , et al
- Makadia R ,
- Matcho A , et al
- Mercadé-Besora N ,
- Kolde R , et al
- Ostropolets A ,
- Makadia R , et al
- Rathod-Mistry T , et al
- Thomas LE ,
- ↵ Coronavirus (COVID-19) in the UK . 2022 . Available : https://coronavirus.data.gov.uk/
- Generalitat de Catalunya
- Schuemie MJ ,
- Hripcsak G ,
- Ryan PB , et al
- Houghton DE ,
- Wysokinski W ,
- Casanegra AI , et al
- Katsoularis I ,
- Fonseca-Rodríguez O ,
- Farrington P , et al
- Jehangir Q ,
- Li P , et al
- Byambasuren O ,
- Stehlik P ,
- Clark J , et al
- Brannock MD ,
- Preiss AJ , et al
- Schneeweiss S , RCT-DUPLICATE Initiative , et al
Supplementary materials
Supplementary data.
This web only file has been produced by the BMJ Publishing Group from an electronic file supplied by the author(s) and has not been edited for content.
- Data supplement 1
AMJ and MC are joint senior authors.
Contributors DPA and AMJ led the conceptualisation of the study with contributions from MC and NM-B. AMJ, TD-S, ER, AU and NTHT adapted the study design with respect to the local vaccine rollouts. AD and WYM mapped and curated CPRD data. MC and NM-B developed code with methodological contributions advice from MTS-S and CP. DPA, MC, NTHT, TD-S, HMEN, XL, CR and AMJ clinically interpreted the results. NM-B, XL, AMJ and DPA wrote the first draft of the manuscript, and all authors read, revised and approved the final version. DPA and AMJ obtained the funding for this research. DPA is responsible for the overall content as guarantor: he accepts full responsibility for the work and the conduct of the study, had access to the data, and controlled the decision to publish.
Funding The research was supported by the National Institute for Health and Care Research (NIHR) Oxford Biomedical Research Centre (BRC). DPA is funded through a NIHR Senior Research Fellowship (Grant number SRF-2018–11-ST2-004). Funding to perform the study in the SIDIAP database was provided by the Real World Epidemiology (RWEpi) research group at IDIAPJGol. Costs of databases mapping to OMOP CDM were covered by the European Health Data and Evidence Network (EHDEN).
Patient and public involvement Patients and/or the public were not involved in the design, or conduct, or reporting or dissemination plans of this research.
Provenance and peer review Not commissioned; externally peer reviewed.
Supplemental material This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Read the full text or download the PDF:
- Download PDF
- Share X Facebook Email LinkedIn
- Permissions
Characteristics of Melatonin Use Among US Children and Adolescents
- 1 Department of Integrative Physiology, University of Colorado Boulder, Boulder
- 2 Department of Public Health, Purdue University, West Lafayette, Indiana
- 3 Sleep Health and Wellness Center, Santa Barbara, California
- 4 Department of Psychiatry and Human Behavior, Alpert Medical School of Brown University, Providence, Rhode Island
In a 2017-2018 study, 1 1.3% of US parents reported that their children consumed melatonin in the past 30 days, and sales more than doubled between 2017 and 2020. 2 In the US, melatonin is considered a dietary supplement, is not regulated by the US Food and Drug Administration, and requires no prescription, raising particular concern because the amount of melatonin present in over-the-counter supplements can vary drastically. In a recent examination of 25 commercial supplements, actual melatonin quantity ranged from 74% to 347% of the labeled content. 3 Additionally, incidence of melatonin ingestion reported to poison control centers increased 530% from 2012 to 2021, 4 largely occurring among children younger than 5 years. Current data are lacking on the prevalence of melatonin use and the frequency, dosing, and timing of melatonin administration in US youth.
Read More About
Hartstein LE , Garrison MM , Lewin D , Boergers J , LeBourgeois MK. Characteristics of Melatonin Use Among US Children and Adolescents. JAMA Pediatr. 2024;178(1):91–93. doi:10.1001/jamapediatrics.2023.4749
Manage citations:
© 2024
Artificial Intelligence Resource Center
Pediatrics in JAMA : Read the Latest
Browse and subscribe to JAMA Network podcasts!
Others Also Liked
Select your interests.
Customize your JAMA Network experience by selecting one or more topics from the list below.
- Academic Medicine
- Acid Base, Electrolytes, Fluids
- Allergy and Clinical Immunology
- American Indian or Alaska Natives
- Anesthesiology
- Anticoagulation
- Art and Images in Psychiatry
- Artificial Intelligence
- Assisted Reproduction
- Bleeding and Transfusion
- Caring for the Critically Ill Patient
- Challenges in Clinical Electrocardiography
- Climate and Health
- Climate Change
- Clinical Challenge
- Clinical Decision Support
- Clinical Implications of Basic Neuroscience
- Clinical Pharmacy and Pharmacology
- Complementary and Alternative Medicine
- Consensus Statements
- Coronavirus (COVID-19)
- Critical Care Medicine
- Cultural Competency
- Dental Medicine
- Dermatology
- Diabetes and Endocrinology
- Diagnostic Test Interpretation
- Drug Development
- Electronic Health Records
- Emergency Medicine
- End of Life, Hospice, Palliative Care
- Environmental Health
- Equity, Diversity, and Inclusion
- Facial Plastic Surgery
- Gastroenterology and Hepatology
- Genetics and Genomics
- Genomics and Precision Health
- Global Health
- Guide to Statistics and Methods
- Hair Disorders
- Health Care Delivery Models
- Health Care Economics, Insurance, Payment
- Health Care Quality
- Health Care Reform
- Health Care Safety
- Health Care Workforce
- Health Disparities
- Health Inequities
- Health Policy
- Health Systems Science
- History of Medicine
- Hypertension
- Images in Neurology
- Implementation Science
- Infectious Diseases
- Innovations in Health Care Delivery
- JAMA Infographic
- Law and Medicine
- Leading Change
- Less is More
- LGBTQIA Medicine
- Lifestyle Behaviors
- Medical Coding
- Medical Devices and Equipment
- Medical Education
- Medical Education and Training
- Medical Journals and Publishing
- Mobile Health and Telemedicine
- Narrative Medicine
- Neuroscience and Psychiatry
- Notable Notes
- Nutrition, Obesity, Exercise
- Obstetrics and Gynecology
- Occupational Health
- Ophthalmology
- Orthopedics
- Otolaryngology
- Pain Medicine
- Palliative Care
- Pathology and Laboratory Medicine
- Patient Care
- Patient Information
- Performance Improvement
- Performance Measures
- Perioperative Care and Consultation
- Pharmacoeconomics
- Pharmacoepidemiology
- Pharmacogenetics
- Pharmacy and Clinical Pharmacology
- Physical Medicine and Rehabilitation
- Physical Therapy
- Physician Leadership
- Population Health
- Primary Care
- Professional Well-being
- Professionalism
- Psychiatry and Behavioral Health
- Public Health
- Pulmonary Medicine
- Regulatory Agencies
- Reproductive Health
- Research, Methods, Statistics
- Resuscitation
- Rheumatology
- Risk Management
- Scientific Discovery and the Future of Medicine
- Shared Decision Making and Communication
- Sleep Medicine
- Sports Medicine
- Stem Cell Transplantation
- Substance Use and Addiction Medicine
- Surgical Innovation
- Surgical Pearls
- Teachable Moment
- Technology and Finance
- The Art of JAMA
- The Arts and Medicine
- The Rational Clinical Examination
- Tobacco and e-Cigarettes
- Translational Medicine
- Trauma and Injury
- Treatment Adherence
- Ultrasonography
- Users' Guide to the Medical Literature
- Vaccination
- Venous Thromboembolism
- Veterans Health
- Women's Health
- Workflow and Process
- Wound Care, Infection, Healing
- Register for email alerts with links to free full-text articles
- Access PDFs of free articles
- Manage your interests
- Save searches and receive search alerts
Thank you for visiting nature.com. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser (or turn off compatibility mode in Internet Explorer). In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript.
- View all journals
- My Account Login
- Explore content
- About the journal
- Publish with us
- Sign up for alerts
- Open access
- Published: 26 March 2024
Predicting and improving complex beer flavor through machine learning
- Michiel Schreurs ORCID: orcid.org/0000-0002-9449-5619 1 , 2 , 3 na1 ,
- Supinya Piampongsant 1 , 2 , 3 na1 ,
- Miguel Roncoroni ORCID: orcid.org/0000-0001-7461-1427 1 , 2 , 3 na1 ,
- Lloyd Cool ORCID: orcid.org/0000-0001-9936-3124 1 , 2 , 3 , 4 ,
- Beatriz Herrera-Malaver ORCID: orcid.org/0000-0002-5096-9974 1 , 2 , 3 ,
- Christophe Vanderaa ORCID: orcid.org/0000-0001-7443-5427 4 ,
- Florian A. Theßeling 1 , 2 , 3 ,
- Łukasz Kreft ORCID: orcid.org/0000-0001-7620-4657 5 ,
- Alexander Botzki ORCID: orcid.org/0000-0001-6691-4233 5 ,
- Philippe Malcorps 6 ,
- Luk Daenen 6 ,
- Tom Wenseleers ORCID: orcid.org/0000-0002-1434-861X 4 &
- Kevin J. Verstrepen ORCID: orcid.org/0000-0002-3077-6219 1 , 2 , 3
Nature Communications volume 15 , Article number: 2368 ( 2024 ) Cite this article
27k Accesses
720 Altmetric
Metrics details
- Chemical engineering
- Gas chromatography
- Machine learning
- Metabolomics
- Taste receptors
The perception and appreciation of food flavor depends on many interacting chemical compounds and external factors, and therefore proves challenging to understand and predict. Here, we combine extensive chemical and sensory analyses of 250 different beers to train machine learning models that allow predicting flavor and consumer appreciation. For each beer, we measure over 200 chemical properties, perform quantitative descriptive sensory analysis with a trained tasting panel and map data from over 180,000 consumer reviews to train 10 different machine learning models. The best-performing algorithm, Gradient Boosting, yields models that significantly outperform predictions based on conventional statistics and accurately predict complex food features and consumer appreciation from chemical profiles. Model dissection allows identifying specific and unexpected compounds as drivers of beer flavor and appreciation. Adding these compounds results in variants of commercial alcoholic and non-alcoholic beers with improved consumer appreciation. Together, our study reveals how big data and machine learning uncover complex links between food chemistry, flavor and consumer perception, and lays the foundation to develop novel, tailored foods with superior flavors.
Similar content being viewed by others
High-speed and large-scale intrinsically stretchable integrated circuits
Donglai Zhong, Can Wu, … Zhenan Bao
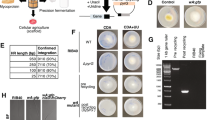
Edible mycelium bioengineered for enhanced nutritional value and sensory appeal using a modular synthetic biology toolkit
Vayu Maini Rekdal, Casper R. B. van der Luijt, … Jay D. Keasling
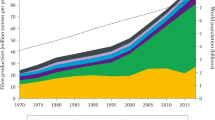
The environmental price of fast fashion
Kirsi Niinimäki, Greg Peters, … Alison Gwilt
Introduction
Predicting and understanding food perception and appreciation is one of the major challenges in food science. Accurate modeling of food flavor and appreciation could yield important opportunities for both producers and consumers, including quality control, product fingerprinting, counterfeit detection, spoilage detection, and the development of new products and product combinations (food pairing) 1 , 2 , 3 , 4 , 5 , 6 . Accurate models for flavor and consumer appreciation would contribute greatly to our scientific understanding of how humans perceive and appreciate flavor. Moreover, accurate predictive models would also facilitate and standardize existing food assessment methods and could supplement or replace assessments by trained and consumer tasting panels, which are variable, expensive and time-consuming 7 , 8 , 9 . Lastly, apart from providing objective, quantitative, accurate and contextual information that can help producers, models can also guide consumers in understanding their personal preferences 10 .
Despite the myriad of applications, predicting food flavor and appreciation from its chemical properties remains a largely elusive goal in sensory science, especially for complex food and beverages 11 , 12 . A key obstacle is the immense number of flavor-active chemicals underlying food flavor. Flavor compounds can vary widely in chemical structure and concentration, making them technically challenging and labor-intensive to quantify, even in the face of innovations in metabolomics, such as non-targeted metabolic fingerprinting 13 , 14 . Moreover, sensory analysis is perhaps even more complicated. Flavor perception is highly complex, resulting from hundreds of different molecules interacting at the physiochemical and sensorial level. Sensory perception is often non-linear, characterized by complex and concentration-dependent synergistic and antagonistic effects 15 , 16 , 17 , 18 , 19 , 20 , 21 that are further convoluted by the genetics, environment, culture and psychology of consumers 22 , 23 , 24 . Perceived flavor is therefore difficult to measure, with problems of sensitivity, accuracy, and reproducibility that can only be resolved by gathering sufficiently large datasets 25 . Trained tasting panels are considered the prime source of quality sensory data, but require meticulous training, are low throughput and high cost. Public databases containing consumer reviews of food products could provide a valuable alternative, especially for studying appreciation scores, which do not require formal training 25 . Public databases offer the advantage of amassing large amounts of data, increasing the statistical power to identify potential drivers of appreciation. However, public datasets suffer from biases, including a bias in the volunteers that contribute to the database, as well as confounding factors such as price, cult status and psychological conformity towards previous ratings of the product.
Classical multivariate statistics and machine learning methods have been used to predict flavor of specific compounds by, for example, linking structural properties of a compound to its potential biological activities or linking concentrations of specific compounds to sensory profiles 1 , 26 . Importantly, most previous studies focused on predicting organoleptic properties of single compounds (often based on their chemical structure) 27 , 28 , 29 , 30 , 31 , 32 , 33 , thus ignoring the fact that these compounds are present in a complex matrix in food or beverages and excluding complex interactions between compounds. Moreover, the classical statistics commonly used in sensory science 34 , 35 , 36 , 37 , 38 , 39 require a large sample size and sufficient variance amongst predictors to create accurate models. They are not fit for studying an extensive set of hundreds of interacting flavor compounds, since they are sensitive to outliers, have a high tendency to overfit and are less suited for non-linear and discontinuous relationships 40 .
In this study, we combine extensive chemical analyses and sensory data of a set of different commercial beers with machine learning approaches to develop models that predict taste, smell, mouthfeel and appreciation from compound concentrations. Beer is particularly suited to model the relationship between chemistry, flavor and appreciation. First, beer is a complex product, consisting of thousands of flavor compounds that partake in complex sensory interactions 41 , 42 , 43 . This chemical diversity arises from the raw materials (malt, yeast, hops, water and spices) and biochemical conversions during the brewing process (kilning, mashing, boiling, fermentation, maturation and aging) 44 , 45 . Second, the advent of the internet saw beer consumers embrace online review platforms, such as RateBeer (ZX Ventures, Anheuser-Busch InBev SA/NV) and BeerAdvocate (Next Glass, inc.). In this way, the beer community provides massive data sets of beer flavor and appreciation scores, creating extraordinarily large sensory databases to complement the analyses of our professional sensory panel. Specifically, we characterize over 200 chemical properties of 250 commercial beers, spread across 22 beer styles, and link these to the descriptive sensory profiling data of a 16-person in-house trained tasting panel and data acquired from over 180,000 public consumer reviews. These unique and extensive datasets enable us to train a suite of machine learning models to predict flavor and appreciation from a beer’s chemical profile. Dissection of the best-performing models allows us to pinpoint specific compounds as potential drivers of beer flavor and appreciation. Follow-up experiments confirm the importance of these compounds and ultimately allow us to significantly improve the flavor and appreciation of selected commercial beers. Together, our study represents a significant step towards understanding complex flavors and reinforces the value of machine learning to develop and refine complex foods. In this way, it represents a stepping stone for further computer-aided food engineering applications 46 .
To generate a comprehensive dataset on beer flavor, we selected 250 commercial Belgian beers across 22 different beer styles (Supplementary Fig. S1 ). Beers with ≤ 4.2% alcohol by volume (ABV) were classified as non-alcoholic and low-alcoholic. Blonds and Tripels constitute a significant portion of the dataset (12.4% and 11.2%, respectively) reflecting their presence on the Belgian beer market and the heterogeneity of beers within these styles. By contrast, lager beers are less diverse and dominated by a handful of brands. Rare styles such as Brut or Faro make up only a small fraction of the dataset (2% and 1%, respectively) because fewer of these beers are produced and because they are dominated by distinct characteristics in terms of flavor and chemical composition.
Extensive analysis identifies relationships between chemical compounds in beer
For each beer, we measured 226 different chemical properties, including common brewing parameters such as alcohol content, iso-alpha acids, pH, sugar concentration 47 , and over 200 flavor compounds (Methods, Supplementary Table S1 ). A large portion (37.2%) are terpenoids arising from hopping, responsible for herbal and fruity flavors 16 , 48 . A second major category are yeast metabolites, such as esters and alcohols, that result in fruity and solvent notes 48 , 49 , 50 . Other measured compounds are primarily derived from malt, or other microbes such as non- Saccharomyces yeasts and bacteria (‘wild flora’). Compounds that arise from spices or staling are labeled under ‘Others’. Five attributes (caloric value, total acids and total ester, hop aroma and sulfur compounds) are calculated from multiple individually measured compounds.
As a first step in identifying relationships between chemical properties, we determined correlations between the concentrations of the compounds (Fig. 1 , upper panel, Supplementary Data 1 and 2 , and Supplementary Fig. S2 . For the sake of clarity, only a subset of the measured compounds is shown in Fig. 1 ). Compounds of the same origin typically show a positive correlation, while absence of correlation hints at parameters varying independently. For example, the hop aroma compounds citronellol, and alpha-terpineol show moderate correlations with each other (Spearman’s rho=0.39 and 0.57), but not with the bittering hop component iso-alpha acids (Spearman’s rho=0.16 and −0.07). This illustrates how brewers can independently modify hop aroma and bitterness by selecting hop varieties and dosage time. If hops are added early in the boiling phase, chemical conversions increase bitterness while aromas evaporate, conversely, late addition of hops preserves aroma but limits bitterness 51 . Similarly, hop-derived iso-alpha acids show a strong anti-correlation with lactic acid and acetic acid, likely reflecting growth inhibition of lactic acid and acetic acid bacteria, or the consequent use of fewer hops in sour beer styles, such as West Flanders ales and Fruit beers, that rely on these bacteria for their distinct flavors 52 . Finally, yeast-derived esters (ethyl acetate, ethyl decanoate, ethyl hexanoate, ethyl octanoate) and alcohols (ethanol, isoamyl alcohol, isobutanol, and glycerol), correlate with Spearman coefficients above 0.5, suggesting that these secondary metabolites are correlated with the yeast genetic background and/or fermentation parameters and may be difficult to influence individually, although the choice of yeast strain may offer some control 53 .

Spearman rank correlations are shown. Descriptors are grouped according to their origin (malt (blue), hops (green), yeast (red), wild flora (yellow), Others (black)), and sensory aspect (aroma, taste, palate, and overall appreciation). Please note that for the chemical compounds, for the sake of clarity, only a subset of the total number of measured compounds is shown, with an emphasis on the key compounds for each source. For more details, see the main text and Methods section. Chemical data can be found in Supplementary Data 1 , correlations between all chemical compounds are depicted in Supplementary Fig. S2 and correlation values can be found in Supplementary Data 2 . See Supplementary Data 4 for sensory panel assessments and Supplementary Data 5 for correlation values between all sensory descriptors.
Interestingly, different beer styles show distinct patterns for some flavor compounds (Supplementary Fig. S3 ). These observations agree with expectations for key beer styles, and serve as a control for our measurements. For instance, Stouts generally show high values for color (darker), while hoppy beers contain elevated levels of iso-alpha acids, compounds associated with bitter hop taste. Acetic and lactic acid are not prevalent in most beers, with notable exceptions such as Kriek, Lambic, Faro, West Flanders ales and Flanders Old Brown, which use acid-producing bacteria ( Lactobacillus and Pediococcus ) or unconventional yeast ( Brettanomyces ) 54 , 55 . Glycerol, ethanol and esters show similar distributions across all beer styles, reflecting their common origin as products of yeast metabolism during fermentation 45 , 53 . Finally, low/no-alcohol beers contain low concentrations of glycerol and esters. This is in line with the production process for most of the low/no-alcohol beers in our dataset, which are produced through limiting fermentation or by stripping away alcohol via evaporation or dialysis, with both methods having the unintended side-effect of reducing the amount of flavor compounds in the final beer 56 , 57 .
Besides expected associations, our data also reveals less trivial associations between beer styles and specific parameters. For example, geraniol and citronellol, two monoterpenoids responsible for citrus, floral and rose flavors and characteristic of Citra hops, are found in relatively high amounts in Christmas, Saison, and Brett/co-fermented beers, where they may originate from terpenoid-rich spices such as coriander seeds instead of hops 58 .
Tasting panel assessments reveal sensorial relationships in beer
To assess the sensory profile of each beer, a trained tasting panel evaluated each of the 250 beers for 50 sensory attributes, including different hop, malt and yeast flavors, off-flavors and spices. Panelists used a tasting sheet (Supplementary Data 3 ) to score the different attributes. Panel consistency was evaluated by repeating 12 samples across different sessions and performing ANOVA. In 95% of cases no significant difference was found across sessions ( p > 0.05), indicating good panel consistency (Supplementary Table S2 ).
Aroma and taste perception reported by the trained panel are often linked (Fig. 1 , bottom left panel and Supplementary Data 4 and 5 ), with high correlations between hops aroma and taste (Spearman’s rho=0.83). Bitter taste was found to correlate with hop aroma and taste in general (Spearman’s rho=0.80 and 0.69), and particularly with “grassy” noble hops (Spearman’s rho=0.75). Barnyard flavor, most often associated with sour beers, is identified together with stale hops (Spearman’s rho=0.97) that are used in these beers. Lactic and acetic acid, which often co-occur, are correlated (Spearman’s rho=0.66). Interestingly, sweetness and bitterness are anti-correlated (Spearman’s rho = −0.48), confirming the hypothesis that they mask each other 59 , 60 . Beer body is highly correlated with alcohol (Spearman’s rho = 0.79), and overall appreciation is found to correlate with multiple aspects that describe beer mouthfeel (alcohol, carbonation; Spearman’s rho= 0.32, 0.39), as well as with hop and ester aroma intensity (Spearman’s rho=0.39 and 0.35).
Similar to the chemical analyses, sensorial analyses confirmed typical features of specific beer styles (Supplementary Fig. S4 ). For example, sour beers (Faro, Flanders Old Brown, Fruit beer, Kriek, Lambic, West Flanders ale) were rated acidic, with flavors of both acetic and lactic acid. Hoppy beers were found to be bitter and showed hop-associated aromas like citrus and tropical fruit. Malt taste is most detected among scotch, stout/porters, and strong ales, while low/no-alcohol beers, which often have a reputation for being ‘worty’ (reminiscent of unfermented, sweet malt extract) appear in the middle. Unsurprisingly, hop aromas are most strongly detected among hoppy beers. Like its chemical counterpart (Supplementary Fig. S3 ), acidity shows a right-skewed distribution, with the most acidic beers being Krieks, Lambics, and West Flanders ales.
Tasting panel assessments of specific flavors correlate with chemical composition
We find that the concentrations of several chemical compounds strongly correlate with specific aroma or taste, as evaluated by the tasting panel (Fig. 2 , Supplementary Fig. S5 , Supplementary Data 6 ). In some cases, these correlations confirm expectations and serve as a useful control for data quality. For example, iso-alpha acids, the bittering compounds in hops, strongly correlate with bitterness (Spearman’s rho=0.68), while ethanol and glycerol correlate with tasters’ perceptions of alcohol and body, the mouthfeel sensation of fullness (Spearman’s rho=0.82/0.62 and 0.72/0.57 respectively) and darker color from roasted malts is a good indication of malt perception (Spearman’s rho=0.54).

Heatmap colors indicate Spearman’s Rho. Axes are organized according to sensory categories (aroma, taste, mouthfeel, overall), chemical categories and chemical sources in beer (malt (blue), hops (green), yeast (red), wild flora (yellow), Others (black)). See Supplementary Data 6 for all correlation values.
Interestingly, for some relationships between chemical compounds and perceived flavor, correlations are weaker than expected. For example, the rose-smelling phenethyl acetate only weakly correlates with floral aroma. This hints at more complex relationships and interactions between compounds and suggests a need for a more complex model than simple correlations. Lastly, we uncovered unexpected correlations. For instance, the esters ethyl decanoate and ethyl octanoate appear to correlate slightly with hop perception and bitterness, possibly due to their fruity flavor. Iron is anti-correlated with hop aromas and bitterness, most likely because it is also anti-correlated with iso-alpha acids. This could be a sign of metal chelation of hop acids 61 , given that our analyses measure unbound hop acids and total iron content, or could result from the higher iron content in dark and Fruit beers, which typically have less hoppy and bitter flavors 62 .
Public consumer reviews complement expert panel data
To complement and expand the sensory data of our trained tasting panel, we collected 180,000 reviews of our 250 beers from the online consumer review platform RateBeer. This provided numerical scores for beer appearance, aroma, taste, palate, overall quality as well as the average overall score.
Public datasets are known to suffer from biases, such as price, cult status and psychological conformity towards previous ratings of a product. For example, prices correlate with appreciation scores for these online consumer reviews (rho=0.49, Supplementary Fig. S6 ), but not for our trained tasting panel (rho=0.19). This suggests that prices affect consumer appreciation, which has been reported in wine 63 , while blind tastings are unaffected. Moreover, we observe that some beer styles, like lagers and non-alcoholic beers, generally receive lower scores, reflecting that online reviewers are mostly beer aficionados with a preference for specialty beers over lager beers. In general, we find a modest correlation between our trained panel’s overall appreciation score and the online consumer appreciation scores (Fig. 3 , rho=0.29). Apart from the aforementioned biases in the online datasets, serving temperature, sample freshness and surroundings, which are all tightly controlled during the tasting panel sessions, can vary tremendously across online consumers and can further contribute to (among others, appreciation) differences between the two categories of tasters. Importantly, in contrast to the overall appreciation scores, for many sensory aspects the results from the professional panel correlated well with results obtained from RateBeer reviews. Correlations were highest for features that are relatively easy to recognize even for untrained tasters, like bitterness, sweetness, alcohol and malt aroma (Fig. 3 and below).
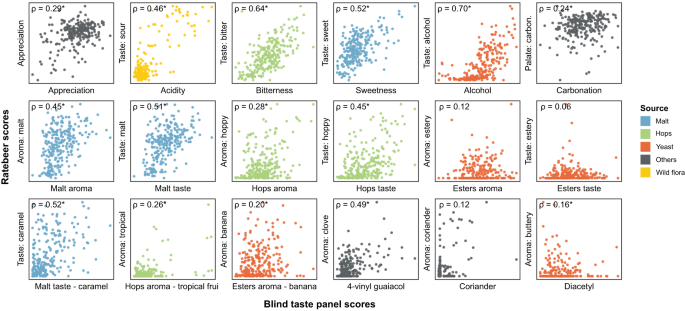
RateBeer text mining results can be found in Supplementary Data 7 . Rho values shown are Spearman correlation values, with asterisks indicating significant correlations ( p < 0.05, two-sided). All p values were smaller than 0.001, except for Esters aroma (0.0553), Esters taste (0.3275), Esters aroma—banana (0.0019), Coriander (0.0508) and Diacetyl (0.0134).
Besides collecting consumer appreciation from these online reviews, we developed automated text analysis tools to gather additional data from review texts (Supplementary Data 7 ). Processing review texts on the RateBeer database yielded comparable results to the scores given by the trained panel for many common sensory aspects, including acidity, bitterness, sweetness, alcohol, malt, and hop tastes (Fig. 3 ). This is in line with what would be expected, since these attributes require less training for accurate assessment and are less influenced by environmental factors such as temperature, serving glass and odors in the environment. Consumer reviews also correlate well with our trained panel for 4-vinyl guaiacol, a compound associated with a very characteristic aroma. By contrast, correlations for more specific aromas like ester, coriander or diacetyl are underrepresented in the online reviews, underscoring the importance of using a trained tasting panel and standardized tasting sheets with explicit factors to be scored for evaluating specific aspects of a beer. Taken together, our results suggest that public reviews are trustworthy for some, but not all, flavor features and can complement or substitute taste panel data for these sensory aspects.
Models can predict beer sensory profiles from chemical data
The rich datasets of chemical analyses, tasting panel assessments and public reviews gathered in the first part of this study provided us with a unique opportunity to develop predictive models that link chemical data to sensorial features. Given the complexity of beer flavor, basic statistical tools such as correlations or linear regression may not always be the most suitable for making accurate predictions. Instead, we applied different machine learning models that can model both simple linear and complex interactive relationships. Specifically, we constructed a set of regression models to predict (a) trained panel scores for beer flavor and quality and (b) public reviews’ appreciation scores from beer chemical profiles. We trained and tested 10 different models (Methods), 3 linear regression-based models (simple linear regression with first-order interactions (LR), lasso regression with first-order interactions (Lasso), partial least squares regressor (PLSR)), 5 decision tree models (AdaBoost regressor (ABR), extra trees (ET), gradient boosting regressor (GBR), random forest (RF) and XGBoost regressor (XGBR)), 1 support vector regression (SVR), and 1 artificial neural network (ANN) model.
To compare the performance of our machine learning models, the dataset was randomly split into a training and test set, stratified by beer style. After a model was trained on data in the training set, its performance was evaluated on its ability to predict the test dataset obtained from multi-output models (based on the coefficient of determination, see Methods). Additionally, individual-attribute models were ranked per descriptor and the average rank was calculated, as proposed by Korneva et al. 64 . Importantly, both ways of evaluating the models’ performance agreed in general. Performance of the different models varied (Table 1 ). It should be noted that all models perform better at predicting RateBeer results than results from our trained tasting panel. One reason could be that sensory data is inherently variable, and this variability is averaged out with the large number of public reviews from RateBeer. Additionally, all tree-based models perform better at predicting taste than aroma. Linear models (LR) performed particularly poorly, with negative R 2 values, due to severe overfitting (training set R 2 = 1). Overfitting is a common issue in linear models with many parameters and limited samples, especially with interaction terms further amplifying the number of parameters. L1 regularization (Lasso) successfully overcomes this overfitting, out-competing multiple tree-based models on the RateBeer dataset. Similarly, the dimensionality reduction of PLSR avoids overfitting and improves performance, to some extent. Still, tree-based models (ABR, ET, GBR, RF and XGBR) show the best performance, out-competing the linear models (LR, Lasso, PLSR) commonly used in sensory science 65 .
GBR models showed the best overall performance in predicting sensory responses from chemical information, with R 2 values up to 0.75 depending on the predicted sensory feature (Supplementary Table S4 ). The GBR models predict consumer appreciation (RateBeer) better than our trained panel’s appreciation (R 2 value of 0.67 compared to R 2 value of 0.09) (Supplementary Table S3 and Supplementary Table S4 ). ANN models showed intermediate performance, likely because neural networks typically perform best with larger datasets 66 . The SVR shows intermediate performance, mostly due to the weak predictions of specific attributes that lower the overall performance (Supplementary Table S4 ).
Model dissection identifies specific, unexpected compounds as drivers of consumer appreciation
Next, we leveraged our models to infer important contributors to sensory perception and consumer appreciation. Consumer preference is a crucial sensory aspects, because a product that shows low consumer appreciation scores often does not succeed commercially 25 . Additionally, the requirement for a large number of representative evaluators makes consumer trials one of the more costly and time-consuming aspects of product development. Hence, a model for predicting chemical drivers of overall appreciation would be a welcome addition to the available toolbox for food development and optimization.
Since GBR models on our RateBeer dataset showed the best overall performance, we focused on these models. Specifically, we used two approaches to identify important contributors. First, rankings of the most important predictors for each sensorial trait in the GBR models were obtained based on impurity-based feature importance (mean decrease in impurity). High-ranked parameters were hypothesized to be either the true causal chemical properties underlying the trait, to correlate with the actual causal properties, or to take part in sensory interactions affecting the trait 67 (Fig. 4A ). In a second approach, we used SHAP 68 to determine which parameters contributed most to the model for making predictions of consumer appreciation (Fig. 4B ). SHAP calculates parameter contributions to model predictions on a per-sample basis, which can be aggregated into an importance score.
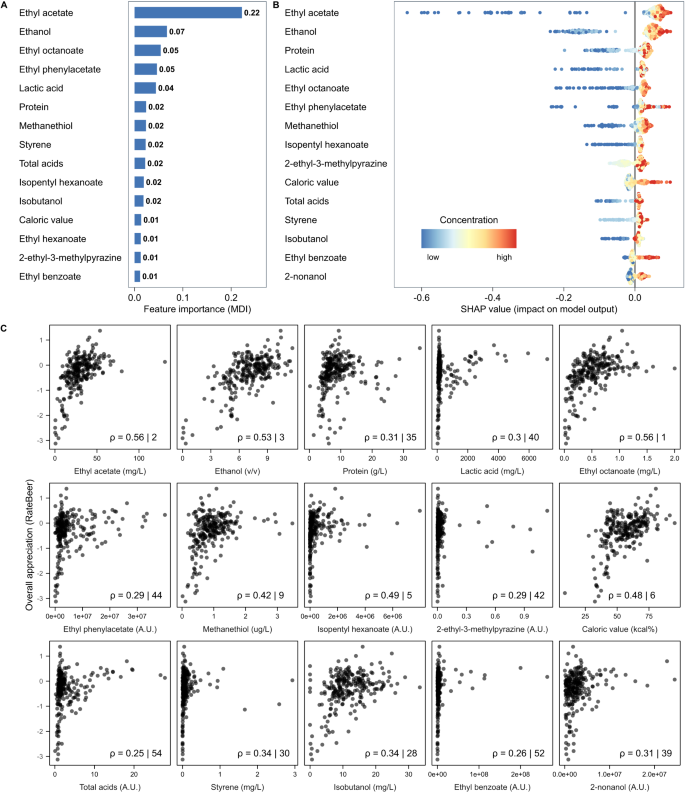
A The impurity-based feature importance (mean deviance in impurity, MDI) calculated from the Gradient Boosting Regression (GBR) model predicting RateBeer appreciation scores. The top 15 highest ranked chemical properties are shown. B SHAP summary plot for the top 15 parameters contributing to our GBR model. Each point on the graph represents a sample from our dataset. The color represents the concentration of that parameter, with bluer colors representing low values and redder colors representing higher values. Greater absolute values on the horizontal axis indicate a higher impact of the parameter on the prediction of the model. C Spearman correlations between the 15 most important chemical properties and consumer overall appreciation. Numbers indicate the Spearman Rho correlation coefficient, and the rank of this correlation compared to all other correlations. The top 15 important compounds were determined using SHAP (panel B).
Both approaches identified ethyl acetate as the most predictive parameter for beer appreciation (Fig. 4 ). Ethyl acetate is the most abundant ester in beer with a typical ‘fruity’, ‘solvent’ and ‘alcoholic’ flavor, but is often considered less important than other esters like isoamyl acetate. The second most important parameter identified by SHAP is ethanol, the most abundant beer compound after water. Apart from directly contributing to beer flavor and mouthfeel, ethanol drastically influences the physical properties of beer, dictating how easily volatile compounds escape the beer matrix to contribute to beer aroma 69 . Importantly, it should also be noted that the importance of ethanol for appreciation is likely inflated by the very low appreciation scores of non-alcoholic beers (Supplementary Fig. S4 ). Despite not often being considered a driver of beer appreciation, protein level also ranks highly in both approaches, possibly due to its effect on mouthfeel and body 70 . Lactic acid, which contributes to the tart taste of sour beers, is the fourth most important parameter identified by SHAP, possibly due to the generally high appreciation of sour beers in our dataset.
Interestingly, some of the most important predictive parameters for our model are not well-established as beer flavors or are even commonly regarded as being negative for beer quality. For example, our models identify methanethiol and ethyl phenyl acetate, an ester commonly linked to beer staling 71 , as a key factor contributing to beer appreciation. Although there is no doubt that high concentrations of these compounds are considered unpleasant, the positive effects of modest concentrations are not yet known 72 , 73 .
To compare our approach to conventional statistics, we evaluated how well the 15 most important SHAP-derived parameters correlate with consumer appreciation (Fig. 4C ). Interestingly, only 6 of the properties derived by SHAP rank amongst the top 15 most correlated parameters. For some chemical compounds, the correlations are so low that they would have likely been considered unimportant. For example, lactic acid, the fourth most important parameter, shows a bimodal distribution for appreciation, with sour beers forming a separate cluster, that is missed entirely by the Spearman correlation. Additionally, the correlation plots reveal outliers, emphasizing the need for robust analysis tools. Together, this highlights the need for alternative models, like the Gradient Boosting model, that better grasp the complexity of (beer) flavor.
Finally, to observe the relationships between these chemical properties and their predicted targets, partial dependence plots were constructed for the six most important predictors of consumer appreciation 74 , 75 , 76 (Supplementary Fig. S7 ). One-way partial dependence plots show how a change in concentration affects the predicted appreciation. These plots reveal an important limitation of our models: appreciation predictions remain constant at ever-increasing concentrations. This implies that once a threshold concentration is reached, further increasing the concentration does not affect appreciation. This is false, as it is well-documented that certain compounds become unpleasant at high concentrations, including ethyl acetate (‘nail polish’) 77 and methanethiol (‘sulfury’ and ‘rotten cabbage’) 78 . The inability of our models to grasp that flavor compounds have optimal levels, above which they become negative, is a consequence of working with commercial beer brands where (off-)flavors are rarely too high to negatively impact the product. The two-way partial dependence plots show how changing the concentration of two compounds influences predicted appreciation, visualizing their interactions (Supplementary Fig. S7 ). In our case, the top 5 parameters are dominated by additive or synergistic interactions, with high concentrations for both compounds resulting in the highest predicted appreciation.
To assess the robustness of our best-performing models and model predictions, we performed 100 iterations of the GBR, RF and ET models. In general, all iterations of the models yielded similar performance (Supplementary Fig. S8 ). Moreover, the main predictors (including the top predictors ethanol and ethyl acetate) remained virtually the same, especially for GBR and RF. For the iterations of the ET model, we did observe more variation in the top predictors, which is likely a consequence of the model’s inherent random architecture in combination with co-correlations between certain predictors. However, even in this case, several of the top predictors (ethanol and ethyl acetate) remain unchanged, although their rank in importance changes (Supplementary Fig. S8 ).
Next, we investigated if a combination of RateBeer and trained panel data into one consolidated dataset would lead to stronger models, under the hypothesis that such a model would suffer less from bias in the datasets. A GBR model was trained to predict appreciation on the combined dataset. This model underperformed compared to the RateBeer model, both in the native case and when including a dataset identifier (R 2 = 0.67, 0.26 and 0.42 respectively). For the latter, the dataset identifier is the most important feature (Supplementary Fig. S9 ), while most of the feature importance remains unchanged, with ethyl acetate and ethanol ranking highest, like in the original model trained only on RateBeer data. It seems that the large variation in the panel dataset introduces noise, weakening the models’ performances and reliability. In addition, it seems reasonable to assume that both datasets are fundamentally different, with the panel dataset obtained by blind tastings by a trained professional panel.
Lastly, we evaluated whether beer style identifiers would further enhance the model’s performance. A GBR model was trained with parameters that explicitly encoded the styles of the samples. This did not improve model performance (R2 = 0.66 with style information vs R2 = 0.67). The most important chemical features are consistent with the model trained without style information (eg. ethanol and ethyl acetate), and with the exception of the most preferred (strong ale) and least preferred (low/no-alcohol) styles, none of the styles were among the most important features (Supplementary Fig. S9 , Supplementary Table S5 and S6 ). This is likely due to a combination of style-specific chemical signatures, such as iso-alpha acids and lactic acid, that implicitly convey style information to the original models, as well as the low number of samples belonging to some styles, making it difficult for the model to learn style-specific patterns. Moreover, beer styles are not rigorously defined, with some styles overlapping in features and some beers being misattributed to a specific style, all of which leads to more noise in models that use style parameters.
Model validation
To test if our predictive models give insight into beer appreciation, we set up experiments aimed at improving existing commercial beers. We specifically selected overall appreciation as the trait to be examined because of its complexity and commercial relevance. Beer flavor comprises a complex bouquet rather than single aromas and tastes 53 . Hence, adding a single compound to the extent that a difference is noticeable may lead to an unbalanced, artificial flavor. Therefore, we evaluated the effect of combinations of compounds. Because Blond beers represent the most extensive style in our dataset, we selected a beer from this style as the starting material for these experiments (Beer 64 in Supplementary Data 1 ).
In the first set of experiments, we adjusted the concentrations of compounds that made up the most important predictors of overall appreciation (ethyl acetate, ethanol, lactic acid, ethyl phenyl acetate) together with correlated compounds (ethyl hexanoate, isoamyl acetate, glycerol), bringing them up to 95 th percentile ethanol-normalized concentrations (Methods) within the Blond group (‘Spiked’ concentration in Fig. 5A ). Compared to controls, the spiked beers were found to have significantly improved overall appreciation among trained panelists, with panelist noting increased intensity of ester flavors, sweetness, alcohol, and body fullness (Fig. 5B ). To disentangle the contribution of ethanol to these results, a second experiment was performed without the addition of ethanol. This resulted in a similar outcome, including increased perception of alcohol and overall appreciation.
Adding the top chemical compounds, identified as best predictors of appreciation by our model, into poorly appreciated beers results in increased appreciation from our trained panel. Results of sensory tests between base beers and those spiked with compounds identified as the best predictors by the model. A Blond and Non/Low-alcohol (0.0% ABV) base beers were brought up to 95th-percentile ethanol-normalized concentrations within each style. B For each sensory attribute, tasters indicated the more intense sample and selected the sample they preferred. The numbers above the bars correspond to the p values that indicate significant changes in perceived flavor (two-sided binomial test: alpha 0.05, n = 20 or 13).
In a last experiment, we tested whether using the model’s predictions can boost the appreciation of a non-alcoholic beer (beer 223 in Supplementary Data 1 ). Again, the addition of a mixture of predicted compounds (omitting ethanol, in this case) resulted in a significant increase in appreciation, body, ester flavor and sweetness.
Predicting flavor and consumer appreciation from chemical composition is one of the ultimate goals of sensory science. A reliable, systematic and unbiased way to link chemical profiles to flavor and food appreciation would be a significant asset to the food and beverage industry. Such tools would substantially aid in quality control and recipe development, offer an efficient and cost-effective alternative to pilot studies and consumer trials and would ultimately allow food manufacturers to produce superior, tailor-made products that better meet the demands of specific consumer groups more efficiently.
A limited set of studies have previously tried, to varying degrees of success, to predict beer flavor and beer popularity based on (a limited set of) chemical compounds and flavors 79 , 80 . Current sensitive, high-throughput technologies allow measuring an unprecedented number of chemical compounds and properties in a large set of samples, yielding a dataset that can train models that help close the gaps between chemistry and flavor, even for a complex natural product like beer. To our knowledge, no previous research gathered data at this scale (250 samples, 226 chemical parameters, 50 sensory attributes and 5 consumer scores) to disentangle and validate the chemical aspects driving beer preference using various machine-learning techniques. We find that modern machine learning models outperform conventional statistical tools, such as correlations and linear models, and can successfully predict flavor appreciation from chemical composition. This could be attributed to the natural incorporation of interactions and non-linear or discontinuous effects in machine learning models, which are not easily grasped by the linear model architecture. While linear models and partial least squares regression represent the most widespread statistical approaches in sensory science, in part because they allow interpretation 65 , 81 , 82 , modern machine learning methods allow for building better predictive models while preserving the possibility to dissect and exploit the underlying patterns. Of the 10 different models we trained, tree-based models, such as our best performing GBR, showed the best overall performance in predicting sensory responses from chemical information, outcompeting artificial neural networks. This agrees with previous reports for models trained on tabular data 83 . Our results are in line with the findings of Colantonio et al. who also identified the gradient boosting architecture as performing best at predicting appreciation and flavor (of tomatoes and blueberries, in their specific study) 26 . Importantly, besides our larger experimental scale, we were able to directly confirm our models’ predictions in vivo.
Our study confirms that flavor compound concentration does not always correlate with perception, suggesting complex interactions that are often missed by more conventional statistics and simple models. Specifically, we find that tree-based algorithms may perform best in developing models that link complex food chemistry with aroma. Furthermore, we show that massive datasets of untrained consumer reviews provide a valuable source of data, that can complement or even replace trained tasting panels, especially for appreciation and basic flavors, such as sweetness and bitterness. This holds despite biases that are known to occur in such datasets, such as price or conformity bias. Moreover, GBR models predict taste better than aroma. This is likely because taste (e.g. bitterness) often directly relates to the corresponding chemical measurements (e.g., iso-alpha acids), whereas such a link is less clear for aromas, which often result from the interplay between multiple volatile compounds. We also find that our models are best at predicting acidity and alcohol, likely because there is a direct relation between the measured chemical compounds (acids and ethanol) and the corresponding perceived sensorial attribute (acidity and alcohol), and because even untrained consumers are generally able to recognize these flavors and aromas.
The predictions of our final models, trained on review data, hold even for blind tastings with small groups of trained tasters, as demonstrated by our ability to validate specific compounds as drivers of beer flavor and appreciation. Since adding a single compound to the extent of a noticeable difference may result in an unbalanced flavor profile, we specifically tested our identified key drivers as a combination of compounds. While this approach does not allow us to validate if a particular single compound would affect flavor and/or appreciation, our experiments do show that this combination of compounds increases consumer appreciation.
It is important to stress that, while it represents an important step forward, our approach still has several major limitations. A key weakness of the GBR model architecture is that amongst co-correlating variables, the largest main effect is consistently preferred for model building. As a result, co-correlating variables often have artificially low importance scores, both for impurity and SHAP-based methods, like we observed in the comparison to the more randomized Extra Trees models. This implies that chemicals identified as key drivers of a specific sensory feature by GBR might not be the true causative compounds, but rather co-correlate with the actual causative chemical. For example, the high importance of ethyl acetate could be (partially) attributed to the total ester content, ethanol or ethyl hexanoate (rho=0.77, rho=0.72 and rho=0.68), while ethyl phenylacetate could hide the importance of prenyl isobutyrate and ethyl benzoate (rho=0.77 and rho=0.76). Expanding our GBR model to include beer style as a parameter did not yield additional power or insight. This is likely due to style-specific chemical signatures, such as iso-alpha acids and lactic acid, that implicitly convey style information to the original model, as well as the smaller sample size per style, limiting the power to uncover style-specific patterns. This can be partly attributed to the curse of dimensionality, where the high number of parameters results in the models mainly incorporating single parameter effects, rather than complex interactions such as style-dependent effects 67 . A larger number of samples may overcome some of these limitations and offer more insight into style-specific effects. On the other hand, beer style is not a rigid scientific classification, and beers within one style often differ a lot, which further complicates the analysis of style as a model factor.
Our study is limited to beers from Belgian breweries. Although these beers cover a large portion of the beer styles available globally, some beer styles and consumer patterns may be missing, while other features might be overrepresented. For example, many Belgian ales exhibit yeast-driven flavor profiles, which is reflected in the chemical drivers of appreciation discovered by this study. In future work, expanding the scope to include diverse markets and beer styles could lead to the identification of even more drivers of appreciation and better models for special niche products that were not present in our beer set.
In addition to inherent limitations of GBR models, there are also some limitations associated with studying food aroma. Even if our chemical analyses measured most of the known aroma compounds, the total number of flavor compounds in complex foods like beer is still larger than the subset we were able to measure in this study. For example, hop-derived thiols, that influence flavor at very low concentrations, are notoriously difficult to measure in a high-throughput experiment. Moreover, consumer perception remains subjective and prone to biases that are difficult to avoid. It is also important to stress that the models are still immature and that more extensive datasets will be crucial for developing more complete models in the future. Besides more samples and parameters, our dataset does not include any demographic information about the tasters. Including such data could lead to better models that grasp external factors like age and culture. Another limitation is that our set of beers consists of high-quality end-products and lacks beers that are unfit for sale, which limits the current model in accurately predicting products that are appreciated very badly. Finally, while models could be readily applied in quality control, their use in sensory science and product development is restrained by their inability to discern causal relationships. Given that the models cannot distinguish compounds that genuinely drive consumer perception from those that merely correlate, validation experiments are essential to identify true causative compounds.
Despite the inherent limitations, dissection of our models enabled us to pinpoint specific molecules as potential drivers of beer aroma and consumer appreciation, including compounds that were unexpected and would not have been identified using standard approaches. Important drivers of beer appreciation uncovered by our models include protein levels, ethyl acetate, ethyl phenyl acetate and lactic acid. Currently, many brewers already use lactic acid to acidify their brewing water and ensure optimal pH for enzymatic activity during the mashing process. Our results suggest that adding lactic acid can also improve beer appreciation, although its individual effect remains to be tested. Interestingly, ethanol appears to be unnecessary to improve beer appreciation, both for blond beer and alcohol-free beer. Given the growing consumer interest in alcohol-free beer, with a predicted annual market growth of >7% 84 , it is relevant for brewers to know what compounds can further increase consumer appreciation of these beers. Hence, our model may readily provide avenues to further improve the flavor and consumer appreciation of both alcoholic and non-alcoholic beers, which is generally considered one of the key challenges for future beer production.
Whereas we see a direct implementation of our results for the development of superior alcohol-free beverages and other food products, our study can also serve as a stepping stone for the development of novel alcohol-containing beverages. We want to echo the growing body of scientific evidence for the negative effects of alcohol consumption, both on the individual level by the mutagenic, teratogenic and carcinogenic effects of ethanol 85 , 86 , as well as the burden on society caused by alcohol abuse and addiction. We encourage the use of our results for the production of healthier, tastier products, including novel and improved beverages with lower alcohol contents. Furthermore, we strongly discourage the use of these technologies to improve the appreciation or addictive properties of harmful substances.
The present work demonstrates that despite some important remaining hurdles, combining the latest developments in chemical analyses, sensory analysis and modern machine learning methods offers exciting avenues for food chemistry and engineering. Soon, these tools may provide solutions in quality control and recipe development, as well as new approaches to sensory science and flavor research.
Beer selection
250 commercial Belgian beers were selected to cover the broad diversity of beer styles and corresponding diversity in chemical composition and aroma. See Supplementary Fig. S1 .
Chemical dataset
Sample preparation.
Beers within their expiration date were purchased from commercial retailers. Samples were prepared in biological duplicates at room temperature, unless explicitly stated otherwise. Bottle pressure was measured with a manual pressure device (Steinfurth Mess-Systeme GmbH) and used to calculate CO 2 concentration. The beer was poured through two filter papers (Macherey-Nagel, 500713032 MN 713 ¼) to remove carbon dioxide and prevent spontaneous foaming. Samples were then prepared for measurements by targeted Headspace-Gas Chromatography-Flame Ionization Detector/Flame Photometric Detector (HS-GC-FID/FPD), Headspace-Solid Phase Microextraction-Gas Chromatography-Mass Spectrometry (HS-SPME-GC-MS), colorimetric analysis, enzymatic analysis, Near-Infrared (NIR) analysis, as described in the sections below. The mean values of biological duplicates are reported for each compound.
HS-GC-FID/FPD
HS-GC-FID/FPD (Shimadzu GC 2010 Plus) was used to measure higher alcohols, acetaldehyde, esters, 4-vinyl guaicol, and sulfur compounds. Each measurement comprised 5 ml of sample pipetted into a 20 ml glass vial containing 1.75 g NaCl (VWR, 27810.295). 100 µl of 2-heptanol (Sigma-Aldrich, H3003) (internal standard) solution in ethanol (Fisher Chemical, E/0650DF/C17) was added for a final concentration of 2.44 mg/L. Samples were flushed with nitrogen for 10 s, sealed with a silicone septum, stored at −80 °C and analyzed in batches of 20.
The GC was equipped with a DB-WAXetr column (length, 30 m; internal diameter, 0.32 mm; layer thickness, 0.50 µm; Agilent Technologies, Santa Clara, CA, USA) to the FID and an HP-5 column (length, 30 m; internal diameter, 0.25 mm; layer thickness, 0.25 µm; Agilent Technologies, Santa Clara, CA, USA) to the FPD. N 2 was used as the carrier gas. Samples were incubated for 20 min at 70 °C in the headspace autosampler (Flow rate, 35 cm/s; Injection volume, 1000 µL; Injection mode, split; Combi PAL autosampler, CTC analytics, Switzerland). The injector, FID and FPD temperatures were kept at 250 °C. The GC oven temperature was first held at 50 °C for 5 min and then allowed to rise to 80 °C at a rate of 5 °C/min, followed by a second ramp of 4 °C/min until 200 °C kept for 3 min and a final ramp of (4 °C/min) until 230 °C for 1 min. Results were analyzed with the GCSolution software version 2.4 (Shimadzu, Kyoto, Japan). The GC was calibrated with a 5% EtOH solution (VWR International) containing the volatiles under study (Supplementary Table S7 ).
HS-SPME-GC-MS
HS-SPME-GC-MS (Shimadzu GCMS-QP-2010 Ultra) was used to measure additional volatile compounds, mainly comprising terpenoids and esters. Samples were analyzed by HS-SPME using a triphase DVB/Carboxen/PDMS 50/30 μm SPME fiber (Supelco Co., Bellefonte, PA, USA) followed by gas chromatography (Thermo Fisher Scientific Trace 1300 series, USA) coupled to a mass spectrometer (Thermo Fisher Scientific ISQ series MS) equipped with a TriPlus RSH autosampler. 5 ml of degassed beer sample was placed in 20 ml vials containing 1.75 g NaCl (VWR, 27810.295). 5 µl internal standard mix was added, containing 2-heptanol (1 g/L) (Sigma-Aldrich, H3003), 4-fluorobenzaldehyde (1 g/L) (Sigma-Aldrich, 128376), 2,3-hexanedione (1 g/L) (Sigma-Aldrich, 144169) and guaiacol (1 g/L) (Sigma-Aldrich, W253200) in ethanol (Fisher Chemical, E/0650DF/C17). Each sample was incubated at 60 °C in the autosampler oven with constant agitation. After 5 min equilibration, the SPME fiber was exposed to the sample headspace for 30 min. The compounds trapped on the fiber were thermally desorbed in the injection port of the chromatograph by heating the fiber for 15 min at 270 °C.
The GC-MS was equipped with a low polarity RXi-5Sil MS column (length, 20 m; internal diameter, 0.18 mm; layer thickness, 0.18 µm; Restek, Bellefonte, PA, USA). Injection was performed in splitless mode at 320 °C, a split flow of 9 ml/min, a purge flow of 5 ml/min and an open valve time of 3 min. To obtain a pulsed injection, a programmed gas flow was used whereby the helium gas flow was set at 2.7 mL/min for 0.1 min, followed by a decrease in flow of 20 ml/min to the normal 0.9 mL/min. The temperature was first held at 30 °C for 3 min and then allowed to rise to 80 °C at a rate of 7 °C/min, followed by a second ramp of 2 °C/min till 125 °C and a final ramp of 8 °C/min with a final temperature of 270 °C.
Mass acquisition range was 33 to 550 amu at a scan rate of 5 scans/s. Electron impact ionization energy was 70 eV. The interface and ion source were kept at 275 °C and 250 °C, respectively. A mix of linear n-alkanes (from C7 to C40, Supelco Co.) was injected into the GC-MS under identical conditions to serve as external retention index markers. Identification and quantification of the compounds were performed using an in-house developed R script as described in Goelen et al. and Reher et al. 87 , 88 (for package information, see Supplementary Table S8 ). Briefly, chromatograms were analyzed using AMDIS (v2.71) 89 to separate overlapping peaks and obtain pure compound spectra. The NIST MS Search software (v2.0 g) in combination with the NIST2017, FFNSC3 and Adams4 libraries were used to manually identify the empirical spectra, taking into account the expected retention time. After background subtraction and correcting for retention time shifts between samples run on different days based on alkane ladders, compound elution profiles were extracted and integrated using a file with 284 target compounds of interest, which were either recovered in our identified AMDIS list of spectra or were known to occur in beer. Compound elution profiles were estimated for every peak in every chromatogram over a time-restricted window using weighted non-negative least square analysis after which peak areas were integrated 87 , 88 . Batch effect correction was performed by normalizing against the most stable internal standard compound, 4-fluorobenzaldehyde. Out of all 284 target compounds that were analyzed, 167 were visually judged to have reliable elution profiles and were used for final analysis.
Discrete photometric and enzymatic analysis
Discrete photometric and enzymatic analysis (Thermo Scientific TM Gallery TM Plus Beermaster Discrete Analyzer) was used to measure acetic acid, ammonia, beta-glucan, iso-alpha acids, color, sugars, glycerol, iron, pH, protein, and sulfite. 2 ml of sample volume was used for the analyses. Information regarding the reagents and standard solutions used for analyses and calibrations is included in Supplementary Table S7 and Supplementary Table S9 .
NIR analyses
NIR analysis (Anton Paar Alcolyzer Beer ME System) was used to measure ethanol. Measurements comprised 50 ml of sample, and a 10% EtOH solution was used for calibration.
Correlation calculations
Pairwise Spearman Rank correlations were calculated between all chemical properties.
Sensory dataset
Trained panel.
Our trained tasting panel consisted of volunteers who gave prior verbal informed consent. All compounds used for the validation experiment were of food-grade quality. The tasting sessions were approved by the Social and Societal Ethics Committee of the KU Leuven (G-2022-5677-R2(MAR)). All online reviewers agreed to the Terms and Conditions of the RateBeer website.
Sensory analysis was performed according to the American Society of Brewing Chemists (ASBC) Sensory Analysis Methods 90 . 30 volunteers were screened through a series of triangle tests. The sixteen most sensitive and consistent tasters were retained as taste panel members. The resulting panel was diverse in age [22–42, mean: 29], sex [56% male] and nationality [7 different countries]. The panel developed a consensus vocabulary to describe beer aroma, taste and mouthfeel. Panelists were trained to identify and score 50 different attributes, using a 7-point scale to rate attributes’ intensity. The scoring sheet is included as Supplementary Data 3 . Sensory assessments took place between 10–12 a.m. The beers were served in black-colored glasses. Per session, between 5 and 12 beers of the same style were tasted at 12 °C to 16 °C. Two reference beers were added to each set and indicated as ‘Reference 1 & 2’, allowing panel members to calibrate their ratings. Not all panelists were present at every tasting. Scores were scaled by standard deviation and mean-centered per taster. Values are represented as z-scores and clustered by Euclidean distance. Pairwise Spearman correlations were calculated between taste and aroma sensory attributes. Panel consistency was evaluated by repeating samples on different sessions and performing ANOVA to identify differences, using the ‘stats’ package (v4.2.2) in R (for package information, see Supplementary Table S8 ).
Online reviews from a public database
The ‘scrapy’ package in Python (v3.6) (for package information, see Supplementary Table S8 ). was used to collect 232,288 online reviews (mean=922, min=6, max=5343) from RateBeer, an online beer review database. Each review entry comprised 5 numerical scores (appearance, aroma, taste, palate and overall quality) and an optional review text. The total number of reviews per reviewer was collected separately. Numerical scores were scaled and centered per rater, and mean scores were calculated per beer.
For the review texts, the language was estimated using the packages ‘langdetect’ and ‘langid’ in Python. Reviews that were classified as English by both packages were kept. Reviewers with fewer than 100 entries overall were discarded. 181,025 reviews from >6000 reviewers from >40 countries remained. Text processing was done using the ‘nltk’ package in Python. Texts were corrected for slang and misspellings; proper nouns and rare words that are relevant to the beer context were specified and kept as-is (‘Chimay’,’Lambic’, etc.). A dictionary of semantically similar sensorial terms, for example ‘floral’ and ‘flower’, was created and collapsed together into one term. Words were stemmed and lemmatized to avoid identifying words such as ‘acid’ and ‘acidity’ as separate terms. Numbers and punctuation were removed.
Sentences from up to 50 randomly chosen reviews per beer were manually categorized according to the aspect of beer they describe (appearance, aroma, taste, palate, overall quality—not to be confused with the 5 numerical scores described above) or flagged as irrelevant if they contained no useful information. If a beer contained fewer than 50 reviews, all reviews were manually classified. This labeled data set was used to train a model that classified the rest of the sentences for all beers 91 . Sentences describing taste and aroma were extracted, and term frequency–inverse document frequency (TFIDF) was implemented to calculate enrichment scores for sensorial words per beer.
The sex of the tasting subject was not considered when building our sensory database. Instead, results from different panelists were averaged, both for our trained panel (56% male, 44% female) and the RateBeer reviews (70% male, 30% female for RateBeer as a whole).
Beer price collection and processing
Beer prices were collected from the following stores: Colruyt, Delhaize, Total Wine, BeerHawk, The Belgian Beer Shop, The Belgian Shop, and Beer of Belgium. Where applicable, prices were converted to Euros and normalized per liter. Spearman correlations were calculated between these prices and mean overall appreciation scores from RateBeer and the taste panel, respectively.
Pairwise Spearman Rank correlations were calculated between all sensory properties.
Machine learning models
Predictive modeling of sensory profiles from chemical data.
Regression models were constructed to predict (a) trained panel scores for beer flavors and quality from beer chemical profiles and (b) public reviews’ appreciation scores from beer chemical profiles. Z-scores were used to represent sensory attributes in both data sets. Chemical properties with log-normal distributions (Shapiro-Wilk test, p < 0.05 ) were log-transformed. Missing chemical measurements (0.1% of all data) were replaced with mean values per attribute. Observations from 250 beers were randomly separated into a training set (70%, 175 beers) and a test set (30%, 75 beers), stratified per beer style. Chemical measurements (p = 231) were normalized based on the training set average and standard deviation. In total, three linear regression-based models: linear regression with first-order interaction terms (LR), lasso regression with first-order interaction terms (Lasso) and partial least squares regression (PLSR); five decision tree models, Adaboost regressor (ABR), Extra Trees (ET), Gradient Boosting regressor (GBR), Random Forest (RF) and XGBoost regressor (XGBR); one support vector machine model (SVR) and one artificial neural network model (ANN) were trained. The models were implemented using the ‘scikit-learn’ package (v1.2.2) and ‘xgboost’ package (v1.7.3) in Python (v3.9.16). Models were trained, and hyperparameters optimized, using five-fold cross-validated grid search with the coefficient of determination (R 2 ) as the evaluation metric. The ANN (scikit-learn’s MLPRegressor) was optimized using Bayesian Tree-Structured Parzen Estimator optimization with the ‘Optuna’ Python package (v3.2.0). Individual models were trained per attribute, and a multi-output model was trained on all attributes simultaneously.
Model dissection
GBR was found to outperform other methods, resulting in models with the highest average R 2 values in both trained panel and public review data sets. Impurity-based rankings of the most important predictors for each predicted sensorial trait were obtained using the ‘scikit-learn’ package. To observe the relationships between these chemical properties and their predicted targets, partial dependence plots (PDP) were constructed for the six most important predictors of consumer appreciation 74 , 75 .
The ‘SHAP’ package in Python (v0.41.0) was implemented to provide an alternative ranking of predictor importance and to visualize the predictors’ effects as a function of their concentration 68 .
Validation of causal chemical properties
To validate the effects of the most important model features on predicted sensory attributes, beers were spiked with the chemical compounds identified by the models and descriptive sensory analyses were carried out according to the American Society of Brewing Chemists (ASBC) protocol 90 .
Compound spiking was done 30 min before tasting. Compounds were spiked into fresh beer bottles, that were immediately resealed and inverted three times. Fresh bottles of beer were opened for the same duration, resealed, and inverted thrice, to serve as controls. Pairs of spiked samples and controls were served simultaneously, chilled and in dark glasses as outlined in the Trained panel section above. Tasters were instructed to select the glass with the higher flavor intensity for each attribute (directional difference test 92 ) and to select the glass they prefer.
The final concentration after spiking was equal to the within-style average, after normalizing by ethanol concentration. This was done to ensure balanced flavor profiles in the final spiked beer. The same methods were applied to improve a non-alcoholic beer. Compounds were the following: ethyl acetate (Merck KGaA, W241415), ethyl hexanoate (Merck KGaA, W243906), isoamyl acetate (Merck KGaA, W205508), phenethyl acetate (Merck KGaA, W285706), ethanol (96%, Colruyt), glycerol (Merck KGaA, W252506), lactic acid (Merck KGaA, 261106).
Significant differences in preference or perceived intensity were determined by performing the two-sided binomial test on each attribute.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The data that support the findings of this work are available in the Supplementary Data files and have been deposited to Zenodo under accession code 10653704 93 . The RateBeer scores data are under restricted access, they are not publicly available as they are property of RateBeer (ZX Ventures, USA). Access can be obtained from the authors upon reasonable request and with permission of RateBeer (ZX Ventures, USA). Source data are provided with this paper.
Code availability
The code for training the machine learning models, analyzing the models, and generating the figures has been deposited to Zenodo under accession code 10653704 93 .
Tieman, D. et al. A chemical genetic roadmap to improved tomato flavor. Science 355 , 391–394 (2017).
Article ADS CAS PubMed Google Scholar
Plutowska, B. & Wardencki, W. Application of gas chromatography–olfactometry (GC–O) in analysis and quality assessment of alcoholic beverages – A review. Food Chem. 107 , 449–463 (2008).
Article CAS Google Scholar
Legin, A., Rudnitskaya, A., Seleznev, B. & Vlasov, Y. Electronic tongue for quality assessment of ethanol, vodka and eau-de-vie. Anal. Chim. Acta 534 , 129–135 (2005).
Loutfi, A., Coradeschi, S., Mani, G. K., Shankar, P. & Rayappan, J. B. B. Electronic noses for food quality: A review. J. Food Eng. 144 , 103–111 (2015).
Ahn, Y.-Y., Ahnert, S. E., Bagrow, J. P. & Barabási, A.-L. Flavor network and the principles of food pairing. Sci. Rep. 1 , 196 (2011).
Article CAS PubMed PubMed Central Google Scholar
Bartoshuk, L. M. & Klee, H. J. Better fruits and vegetables through sensory analysis. Curr. Biol. 23 , R374–R378 (2013).
Article CAS PubMed Google Scholar
Piggott, J. R. Design questions in sensory and consumer science. Food Qual. Prefer. 3293 , 217–220 (1995).
Article Google Scholar
Kermit, M. & Lengard, V. Assessing the performance of a sensory panel-panellist monitoring and tracking. J. Chemom. 19 , 154–161 (2005).
Cook, D. J., Hollowood, T. A., Linforth, R. S. T. & Taylor, A. J. Correlating instrumental measurements of texture and flavour release with human perception. Int. J. Food Sci. Technol. 40 , 631–641 (2005).
Chinchanachokchai, S., Thontirawong, P. & Chinchanachokchai, P. A tale of two recommender systems: The moderating role of consumer expertise on artificial intelligence based product recommendations. J. Retail. Consum. Serv. 61 , 1–12 (2021).
Ross, C. F. Sensory science at the human-machine interface. Trends Food Sci. Technol. 20 , 63–72 (2009).
Chambers, E. IV & Koppel, K. Associations of volatile compounds with sensory aroma and flavor: The complex nature of flavor. Molecules 18 , 4887–4905 (2013).
Pinu, F. R. Metabolomics—The new frontier in food safety and quality research. Food Res. Int. 72 , 80–81 (2015).
Danezis, G. P., Tsagkaris, A. S., Brusic, V. & Georgiou, C. A. Food authentication: state of the art and prospects. Curr. Opin. Food Sci. 10 , 22–31 (2016).
Shepherd, G. M. Smell images and the flavour system in the human brain. Nature 444 , 316–321 (2006).
Meilgaard, M. C. Prediction of flavor differences between beers from their chemical composition. J. Agric. Food Chem. 30 , 1009–1017 (1982).
Xu, L. et al. Widespread receptor-driven modulation in peripheral olfactory coding. Science 368 , eaaz5390 (2020).
Kupferschmidt, K. Following the flavor. Science 340 , 808–809 (2013).
Billesbølle, C. B. et al. Structural basis of odorant recognition by a human odorant receptor. Nature 615 , 742–749 (2023).
Article ADS PubMed PubMed Central Google Scholar
Smith, B. Perspective: Complexities of flavour. Nature 486 , S6–S6 (2012).
Pfister, P. et al. Odorant receptor inhibition is fundamental to odor encoding. Curr. Biol. 30 , 2574–2587 (2020).
Moskowitz, H. W., Kumaraiah, V., Sharma, K. N., Jacobs, H. L. & Sharma, S. D. Cross-cultural differences in simple taste preferences. Science 190 , 1217–1218 (1975).
Eriksson, N. et al. A genetic variant near olfactory receptor genes influences cilantro preference. Flavour 1 , 22 (2012).
Ferdenzi, C. et al. Variability of affective responses to odors: Culture, gender, and olfactory knowledge. Chem. Senses 38 , 175–186 (2013).
Article PubMed Google Scholar
Lawless, H. T. & Heymann, H. Sensory evaluation of food: Principles and practices. (Springer, New York, NY). https://doi.org/10.1007/978-1-4419-6488-5 (2010).
Colantonio, V. et al. Metabolomic selection for enhanced fruit flavor. Proc. Natl. Acad. Sci. 119 , e2115865119 (2022).
Fritz, F., Preissner, R. & Banerjee, P. VirtualTaste: a web server for the prediction of organoleptic properties of chemical compounds. Nucleic Acids Res 49 , W679–W684 (2021).
Tuwani, R., Wadhwa, S. & Bagler, G. BitterSweet: Building machine learning models for predicting the bitter and sweet taste of small molecules. Sci. Rep. 9 , 1–13 (2019).
Dagan-Wiener, A. et al. Bitter or not? BitterPredict, a tool for predicting taste from chemical structure. Sci. Rep. 7 , 1–13 (2017).
Pallante, L. et al. Toward a general and interpretable umami taste predictor using a multi-objective machine learning approach. Sci. Rep. 12 , 1–11 (2022).
Malavolta, M. et al. A survey on computational taste predictors. Eur. Food Res. Technol. 248 , 2215–2235 (2022).
Lee, B. K. et al. A principal odor map unifies diverse tasks in olfactory perception. Science 381 , 999–1006 (2023).
Mayhew, E. J. et al. Transport features predict if a molecule is odorous. Proc. Natl. Acad. Sci. 119 , e2116576119 (2022).
Niu, Y. et al. Sensory evaluation of the synergism among ester odorants in light aroma-type liquor by odor threshold, aroma intensity and flash GC electronic nose. Food Res. Int. 113 , 102–114 (2018).
Yu, P., Low, M. Y. & Zhou, W. Design of experiments and regression modelling in food flavour and sensory analysis: A review. Trends Food Sci. Technol. 71 , 202–215 (2018).
Oladokun, O. et al. The impact of hop bitter acid and polyphenol profiles on the perceived bitterness of beer. Food Chem. 205 , 212–220 (2016).
Linforth, R., Cabannes, M., Hewson, L., Yang, N. & Taylor, A. Effect of fat content on flavor delivery during consumption: An in vivo model. J. Agric. Food Chem. 58 , 6905–6911 (2010).
Guo, S., Na Jom, K. & Ge, Y. Influence of roasting condition on flavor profile of sunflower seeds: A flavoromics approach. Sci. Rep. 9 , 11295 (2019).
Ren, Q. et al. The changes of microbial community and flavor compound in the fermentation process of Chinese rice wine using Fagopyrum tataricum grain as feedstock. Sci. Rep. 9 , 3365 (2019).
Hastie, T., Friedman, J. & Tibshirani, R. The Elements of Statistical Learning. (Springer, New York, NY). https://doi.org/10.1007/978-0-387-21606-5 (2001).
Dietz, C., Cook, D., Huismann, M., Wilson, C. & Ford, R. The multisensory perception of hop essential oil: a review. J. Inst. Brew. 126 , 320–342 (2020).
CAS Google Scholar
Roncoroni, Miguel & Verstrepen, Kevin Joan. Belgian Beer: Tested and Tasted. (Lannoo, 2018).
Meilgaard, M. Flavor chemistry of beer: Part II: Flavor and threshold of 239 aroma volatiles. in (1975).
Bokulich, N. A. & Bamforth, C. W. The microbiology of malting and brewing. Microbiol. Mol. Biol. Rev. MMBR 77 , 157–172 (2013).
Dzialo, M. C., Park, R., Steensels, J., Lievens, B. & Verstrepen, K. J. Physiology, ecology and industrial applications of aroma formation in yeast. FEMS Microbiol. Rev. 41 , S95–S128 (2017).
Article PubMed PubMed Central Google Scholar
Datta, A. et al. Computer-aided food engineering. Nat. Food 3 , 894–904 (2022).
American Society of Brewing Chemists. Beer Methods. (American Society of Brewing Chemists, St. Paul, MN, U.S.A.).
Olaniran, A. O., Hiralal, L., Mokoena, M. P. & Pillay, B. Flavour-active volatile compounds in beer: production, regulation and control. J. Inst. Brew. 123 , 13–23 (2017).
Verstrepen, K. J. et al. Flavor-active esters: Adding fruitiness to beer. J. Biosci. Bioeng. 96 , 110–118 (2003).
Meilgaard, M. C. Flavour chemistry of beer. part I: flavour interaction between principal volatiles. Master Brew. Assoc. Am. Tech. Q 12 , 107–117 (1975).
Briggs, D. E., Boulton, C. A., Brookes, P. A. & Stevens, R. Brewing 227–254. (Woodhead Publishing). https://doi.org/10.1533/9781855739062.227 (2004).
Bossaert, S., Crauwels, S., De Rouck, G. & Lievens, B. The power of sour - A review: Old traditions, new opportunities. BrewingScience 72 , 78–88 (2019).
Google Scholar
Verstrepen, K. J. et al. Flavor active esters: Adding fruitiness to beer. J. Biosci. Bioeng. 96 , 110–118 (2003).
Snauwaert, I. et al. Microbial diversity and metabolite composition of Belgian red-brown acidic ales. Int. J. Food Microbiol. 221 , 1–11 (2016).
Spitaels, F. et al. The microbial diversity of traditional spontaneously fermented lambic beer. PLoS ONE 9 , e95384 (2014).
Blanco, C. A., Andrés-Iglesias, C. & Montero, O. Low-alcohol Beers: Flavor Compounds, Defects, and Improvement Strategies. Crit. Rev. Food Sci. Nutr. 56 , 1379–1388 (2016).
Jackowski, M. & Trusek, A. Non-Alcohol. beer Prod. – Overv. 20 , 32–38 (2018).
Takoi, K. et al. The contribution of geraniol metabolism to the citrus flavour of beer: Synergy of geraniol and β-citronellol under coexistence with excess linalool. J. Inst. Brew. 116 , 251–260 (2010).
Kroeze, J. H. & Bartoshuk, L. M. Bitterness suppression as revealed by split-tongue taste stimulation in humans. Physiol. Behav. 35 , 779–783 (1985).
Mennella, J. A. et al. A spoonful of sugar helps the medicine go down”: Bitter masking bysucrose among children and adults. Chem. Senses 40 , 17–25 (2015).
Wietstock, P., Kunz, T., Perreira, F. & Methner, F.-J. Metal chelation behavior of hop acids in buffered model systems. BrewingScience 69 , 56–63 (2016).
Sancho, D., Blanco, C. A., Caballero, I. & Pascual, A. Free iron in pale, dark and alcohol-free commercial lager beers. J. Sci. Food Agric. 91 , 1142–1147 (2011).
Rodrigues, H. & Parr, W. V. Contribution of cross-cultural studies to understanding wine appreciation: A review. Food Res. Int. 115 , 251–258 (2019).
Korneva, E. & Blockeel, H. Towards better evaluation of multi-target regression models. in ECML PKDD 2020 Workshops (eds. Koprinska, I. et al.) 353–362 (Springer International Publishing, Cham, 2020). https://doi.org/10.1007/978-3-030-65965-3_23 .
Gastón Ares. Mathematical and Statistical Methods in Food Science and Technology. (Wiley, 2013).
Grinsztajn, L., Oyallon, E. & Varoquaux, G. Why do tree-based models still outperform deep learning on tabular data? Preprint at http://arxiv.org/abs/2207.08815 (2022).
Gries, S. T. Statistics for Linguistics with R: A Practical Introduction. in Statistics for Linguistics with R (De Gruyter Mouton, 2021). https://doi.org/10.1515/9783110718256 .
Lundberg, S. M. et al. From local explanations to global understanding with explainable AI for trees. Nat. Mach. Intell. 2 , 56–67 (2020).
Ickes, C. M. & Cadwallader, K. R. Effects of ethanol on flavor perception in alcoholic beverages. Chemosens. Percept. 10 , 119–134 (2017).
Kato, M. et al. Influence of high molecular weight polypeptides on the mouthfeel of commercial beer. J. Inst. Brew. 127 , 27–40 (2021).
Wauters, R. et al. Novel Saccharomyces cerevisiae variants slow down the accumulation of staling aldehydes and improve beer shelf-life. Food Chem. 398 , 1–11 (2023).
Li, H., Jia, S. & Zhang, W. Rapid determination of low-level sulfur compounds in beer by headspace gas chromatography with a pulsed flame photometric detector. J. Am. Soc. Brew. Chem. 66 , 188–191 (2008).
Dercksen, A., Laurens, J., Torline, P., Axcell, B. C. & Rohwer, E. Quantitative analysis of volatile sulfur compounds in beer using a membrane extraction interface. J. Am. Soc. Brew. Chem. 54 , 228–233 (1996).
Molnar, C. Interpretable Machine Learning: A Guide for Making Black-Box Models Interpretable. (2020).
Zhao, Q. & Hastie, T. Causal interpretations of black-box models. J. Bus. Econ. Stat. Publ. Am. Stat. Assoc. 39 , 272–281 (2019).
Article MathSciNet Google Scholar
Hastie, T., Tibshirani, R. & Friedman, J. The Elements of Statistical Learning. (Springer, 2019).
Labrado, D. et al. Identification by NMR of key compounds present in beer distillates and residual phases after dealcoholization by vacuum distillation. J. Sci. Food Agric. 100 , 3971–3978 (2020).
Lusk, L. T., Kay, S. B., Porubcan, A. & Ryder, D. S. Key olfactory cues for beer oxidation. J. Am. Soc. Brew. Chem. 70 , 257–261 (2012).
Gonzalez Viejo, C., Torrico, D. D., Dunshea, F. R. & Fuentes, S. Development of artificial neural network models to assess beer acceptability based on sensory properties using a robotic pourer: A comparative model approach to achieve an artificial intelligence system. Beverages 5 , 33 (2019).
Gonzalez Viejo, C., Fuentes, S., Torrico, D. D., Godbole, A. & Dunshea, F. R. Chemical characterization of aromas in beer and their effect on consumers liking. Food Chem. 293 , 479–485 (2019).
Gilbert, J. L. et al. Identifying breeding priorities for blueberry flavor using biochemical, sensory, and genotype by environment analyses. PLOS ONE 10 , 1–21 (2015).
Goulet, C. et al. Role of an esterase in flavor volatile variation within the tomato clade. Proc. Natl. Acad. Sci. 109 , 19009–19014 (2012).
Article ADS CAS PubMed PubMed Central Google Scholar
Borisov, V. et al. Deep Neural Networks and Tabular Data: A Survey. IEEE Trans. Neural Netw. Learn. Syst. 1–21 https://doi.org/10.1109/TNNLS.2022.3229161 (2022).
Statista. Statista Consumer Market Outlook: Beer - Worldwide.
Seitz, H. K. & Stickel, F. Molecular mechanisms of alcoholmediated carcinogenesis. Nat. Rev. Cancer 7 , 599–612 (2007).
Voordeckers, K. et al. Ethanol exposure increases mutation rate through error-prone polymerases. Nat. Commun. 11 , 3664 (2020).
Goelen, T. et al. Bacterial phylogeny predicts volatile organic compound composition and olfactory response of an aphid parasitoid. Oikos 129 , 1415–1428 (2020).
Article ADS Google Scholar
Reher, T. et al. Evaluation of hop (Humulus lupulus) as a repellent for the management of Drosophila suzukii. Crop Prot. 124 , 104839 (2019).
Stein, S. E. An integrated method for spectrum extraction and compound identification from gas chromatography/mass spectrometry data. J. Am. Soc. Mass Spectrom. 10 , 770–781 (1999).
American Society of Brewing Chemists. Sensory Analysis Methods. (American Society of Brewing Chemists, St. Paul, MN, U.S.A., 1992).
McAuley, J., Leskovec, J. & Jurafsky, D. Learning Attitudes and Attributes from Multi-Aspect Reviews. Preprint at https://doi.org/10.48550/arXiv.1210.3926 (2012).
Meilgaard, M. C., Carr, B. T. & Carr, B. T. Sensory Evaluation Techniques. (CRC Press, Boca Raton). https://doi.org/10.1201/b16452 (2014).
Schreurs, M. et al. Data from: Predicting and improving complex beer flavor through machine learning. Zenodo https://doi.org/10.5281/zenodo.10653704 (2024).
Download references
Acknowledgements
We thank all lab members for their discussions and thank all tasting panel members for their contributions. Special thanks go out to Dr. Karin Voordeckers for her tremendous help in proofreading and improving the manuscript. M.S. was supported by a Baillet-Latour fellowship, L.C. acknowledges financial support from KU Leuven (C16/17/006), F.A.T. was supported by a PhD fellowship from FWO (1S08821N). Research in the lab of K.J.V. is supported by KU Leuven, FWO, VIB, VLAIO and the Brewing Science Serves Health Fund. Research in the lab of T.W. is supported by FWO (G.0A51.15) and KU Leuven (C16/17/006).
Author information
These authors contributed equally: Michiel Schreurs, Supinya Piampongsant, Miguel Roncoroni.
Authors and Affiliations
VIB—KU Leuven Center for Microbiology, Gaston Geenslaan 1, B-3001, Leuven, Belgium
Michiel Schreurs, Supinya Piampongsant, Miguel Roncoroni, Lloyd Cool, Beatriz Herrera-Malaver, Florian A. Theßeling & Kevin J. Verstrepen
CMPG Laboratory of Genetics and Genomics, KU Leuven, Gaston Geenslaan 1, B-3001, Leuven, Belgium
Leuven Institute for Beer Research (LIBR), Gaston Geenslaan 1, B-3001, Leuven, Belgium
Laboratory of Socioecology and Social Evolution, KU Leuven, Naamsestraat 59, B-3000, Leuven, Belgium
Lloyd Cool, Christophe Vanderaa & Tom Wenseleers
VIB Bioinformatics Core, VIB, Rijvisschestraat 120, B-9052, Ghent, Belgium
Łukasz Kreft & Alexander Botzki
AB InBev SA/NV, Brouwerijplein 1, B-3000, Leuven, Belgium
Philippe Malcorps & Luk Daenen
You can also search for this author in PubMed Google Scholar
Contributions
S.P., M.S. and K.J.V. conceived the experiments. S.P., M.S. and K.J.V. designed the experiments. S.P., M.S., M.R., B.H. and F.A.T. performed the experiments. S.P., M.S., L.C., C.V., L.K., A.B., P.M., L.D., T.W. and K.J.V. contributed analysis ideas. S.P., M.S., L.C., C.V., T.W. and K.J.V. analyzed the data. All authors contributed to writing the manuscript.
Corresponding author
Correspondence to Kevin J. Verstrepen .
Ethics declarations
Competing interests.
K.J.V. is affiliated with bar.on. The other authors declare no competing interests.
Peer review
Peer review information.
Nature Communications thanks Florian Bauer, Andrew John Macintosh and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information, peer review file, description of additional supplementary files, supplementary data 1, supplementary data 2, supplementary data 3, supplementary data 4, supplementary data 5, supplementary data 6, supplementary data 7, reporting summary, source data, source data, rights and permissions.
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/ .
Reprints and permissions
About this article
Cite this article.
Schreurs, M., Piampongsant, S., Roncoroni, M. et al. Predicting and improving complex beer flavor through machine learning. Nat Commun 15 , 2368 (2024). https://doi.org/10.1038/s41467-024-46346-0
Download citation
Received : 30 October 2023
Accepted : 21 February 2024
Published : 26 March 2024
DOI : https://doi.org/10.1038/s41467-024-46346-0
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
By submitting a comment you agree to abide by our Terms and Community Guidelines . If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.
Quick links
- Explore articles by subject
- Guide to authors
- Editorial policies
Sign up for the Nature Briefing: Translational Research newsletter — top stories in biotechnology, drug discovery and pharma.

VIDEO
COMMENTS
This article provides an overview of writing for publication in peer-reviewed journals. While the main focus is on writing a research article, it also provides guidance on factors influencing journal selection, including journal scope, intended audience for the findings, open access requirements, and journal citation metrics.
Introduction. This guide provides an overview of the process of preparing and submitting a scholarly manuscript for publication in a psychology journal. Drawing on the experiences of authors of scholarly writings, peer reviewers, and journal editors, we seek to demystify the publication process and to offer advice designed to improve a ...
Report results fully & honestly, as pre-specified. Text (story), Tables (evidence), Figures (highlights) Report primary outcomes first. Give confidence intervals for main results. Report essential summary statistics. Leave out non-essential tables and figures; these can be included as supplementary files. Don't start discussion here.
Paragraph 1: Summarize the Findings. The first paragraph of the discussion should be used to summarize the 1 or 2 key findings from the study. You've taken the reader on a long journey so far, so this is a good time to "refresh" in plain language what this study was about and what the key findings were.
We describe here the basic steps to follow in writing a scientific article. We outline the main sections that an average article should contain; the elements that should appear in these sections, and some pointers for making the overall result attractive and acceptable for publication. 1.
3 or 4 data sets per figure; well-selected scales; appropriate axis label size; symbols clear to read; data sets easily distinguishable. Each photograph must have a scale marker of professional quality in a corner. Use color ONLY when necessary. Color must be visible and distinguishable when printed in black & white.
Look at examples from your target journal to decide the appropriate length. This section should include the elements shown in Fig. 1. Begin with a general context, narrowing to the specific focus of the pa-per. Include five main elements: why your research is im-portant, what is already known about the topic, the gap.
Writing research papers does not come naturally to most of us. The typical research paper is a highly codified rhetorical form [1, 2]. Knowledge of the rules—some explicit, others implied—goes a long way toward writing a paper that will get accepted in a peer-reviewed journal. Primacy of the research question
Rule 1: Focus your paper on a central contribution, which you communicate in the title. Rule 2: Write for flesh-and-blood human beings who do not know your work. Rule 3: Stick to the context-content-conclusion (C-C-C) scheme. Rule 4: Optimise your logical flow by avoiding zig-zag and using parallelism.
The introduction section should be approximately three to five paragraphs in length. Look at examples from your target journal to decide the appropriate length. This section should include the elements shown in Fig. 1. Begin with a general context, narrowing to the specific focus of the paper.
Abstract. Scientific writing and publication are essential to advancing knowledge and practice in public health, but prospective authors face substantial challenges. Authors can overcome barriers, such as lack of understanding about scientific writing and the publishing process, with training and resources. The objective of this article is to ...
Research paper Writing a scientific article: A step-by-step guide for beginners F. Ecarnot*, M.-F. Seronde, R. Chopard, F. Schiele, N. Meneveau ... publications Writing Research Article A B S T R A C T Many young researchers find it extremely difficult to write scientific articles, and few receive specific ...
There is no 'generic' good article that could fit in any journal. Before you start writing, then, the most important task is to choose the journal. You have the research that you have been working on for several years. Setting, data, conclusions, etc. will be the same in almost every paper you are going to produce.
Today publishing articles is a trend around the world almost in each university. Millions of research articles are published in thousands of journals annually throughout many streams/sectors such as medical, engineering, science, etc. But few researchers follow the proper and fundamental criteria to write a quality research article.
readership varies. A proper choice of journal can make a larger impact of your research. Get to know the focus and readership of the journal that you are considering. - general vs. specialized area journal Select 2 or 3 journals in the chosen area with relatively high impact factors. Discuss with your advisor and decide on the journal
There are several different types of article; each journal will have its preferred types and may not accept others. The most common types of article are: l Evidence synthesis articles (such as systematic review articles) l Original research articles l whose first language is not English. Sometimes it might be Clinical articles l Discussion articles
Once this is all completed, the article can be formally submitted (usually via email or an online submission system). Figure 2 provides a sample process for a manuscript once submitted to a journal for consideration for publication. Figure 2: Sample process for a submitted manuscript. Source: The Pharmaceutical Journal.
The present paper discusses about the techniques of writing good research article. The author has traced the procedures and rule of making abstract of an article. The steps needed to make a good ...
State why the problem you address is important. State what is lacking in the current knowledge. State the objectives of your study or the research question. Methods. Describe the context and setting of the study. Specify the study design. Describe the 'population' (patients, doctors, hospitals, etc.) Describe the sampling strategy.
Writing a journal article can be an overwhelming process, but breaking it down into manageable tasks can make the overwhelming the routine. These manageable tasks can be identified by determining what the essential elements of a successful article are and how they function together to produce the desired result: a published journal article.
B ACKGROUND. The publication of original research in a peer-reviewed and indexed journal is the ultimate and most important step toward the recognition of any scientific work.However, the process starts long before the write-up of a manuscript. The journal in which the author wishes to publish his/her work should be chosen at the time of conceptualization of the scientific work based on the ...
Hence, try to find out those reasons and then elaborate that in detail. [1] Do not repeat what has already been stated in the introduction. Before writing discussion, sort out the important ...
A.E. Bender. The essential requirements of a scientific paper are described although each journal publishes its own specific 'Guidelines for Contributors' which may differ to some extent from ...
One study reported data from both routine clinical practice and clinical research. 3 For the purposes of subsequent analysis, this study was split into research and clinical subsamples (therefore, 13 samples are considered henceforth). Antipsychotic status at the time of neuroimaging was reported in 6 samples (n = 714).
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self ...
Request full-text PDF. To read the full-text of this research, you can request a copy directly from the author. ... English for Research Publication Purposes (ERPP) writing and have useful ...
Objective To study the association between COVID-19 vaccination and the risk of post-COVID-19 cardiac and thromboembolic complications. Methods We conducted a staggered cohort study based on national vaccination campaigns using electronic health records from the UK, Spain and Estonia. Vaccine rollout was grouped into four stages with predefined enrolment periods. Each stage included all ...
In a 2017-2018 study, 1 1.3% of US parents reported that their children consumed melatonin in the past 30 days, and sales more than doubled between 2017 and 2020. 2 In the US, melatonin is considered a dietary supplement, is not regulated by the US Food and Drug Administration, and requires no prescription, raising particular concern because the amount of melatonin present in over-the-counter ...
Request full-text PDF. To read the full-text of this research, you can request a copy directly from the authors.
Interestingly, different beer styles show distinct patterns for some flavor compounds (Supplementary Fig. S3).These observations agree with expectations for key beer styles, and serve as a control ...