Thank you for visiting nature.com. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser (or turn off compatibility mode in Internet Explorer). In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript.
- View all journals

Microbiology articles from across Nature Portfolio
Microbiology is the study of microscopic organisms, such as bacteria, viruses, archaea, fungi and protozoa. This discipline includes fundamental research on the biochemistry, physiology, cell biology, ecology, evolution and clinical aspects of microorganisms, including the host response to these agents.
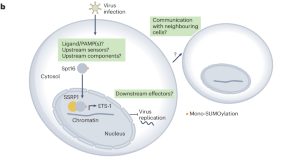
Primal FEAR protects against infection
FEAR is an ancestral histone chaperone complex that can control virus infections in an interferon-independent manner.
- Derek Walsh
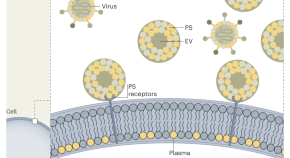
Extracellular vesicles block viral entryways
Extracellular vesicles carrying phosphatidylserine on their surface, found in large quantities in semen, saliva and breast milk, but not in blood, provide an innate defence strategy by blocking viral entry through competition for binding to cellular phosphatidylserine receptors, explaining why many viruses are transmitted by blood rather than by these body fluids.
- Leonid Margolis
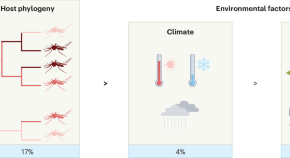
Host species drive composition of mosquito virome
Metatranscriptomic data from more than 2,000 mosquitoes of 81 species show that the composition of mosquito viral communities is determined more by host phylogeny than by climate and land-use factors, which will help to inform arbovirus surveillance.
- Sarah François
Related Subjects
- Antimicrobials
- Applied microbiology
- Bacteriology
- Bacteriophages
- Biogeochemistry
- Cellular microbiology
- Clinical microbiology
- Microbial communities
- CRISPR-Cas systems
- Environmental microbiology
- Industrial microbiology
- Infectious-disease diagnostics
- Microbial genetics
- Parasitology
- Phage biology
- Policy and public health in microbiology
Latest Research and Reviews

Microbial composition associated with biliary stents in patients undergoing pancreatic resection for cancer
- Aitor Blanco-Míguez
- Sara Carloni
- Nicola Segata
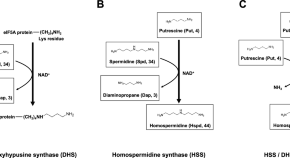
Caldomycin, a new guanidopolyamine produced by a novel agmatine homocoupling enzyme involved in homospermidine biosynthesis
- Teruyuki Kobayashi
- Akihiko Sakamoto
- Yusuke Terui
Coconut rhinoceros beetle digestive symbiosis with potential plant cell wall degrading microbes
- Chiao-Jung Han
- Chih-Hsin Cheng
- Matan Shelomi
Mitochondrial injury induced by a Salmonella genotoxin triggers the proinflammatory senescence-associated secretory phenotype
Chen et al. probe the role of a genotoxin of Salmonella typhi in triggering a senescence-associated secretory phenotype, via mitochondrial DNA damage.
- Han-Yi Chen
- Wan-Chen Hsieh
- Shu-Jung Chang
Colitis reduces active social engagement in mice and is ameliorated by supplementation with human microbiota members
Past intestinal distress is associated with diminished social behavior in mice. Here, the authors show that treatment with microbiota members that are enriched in neurotypical people versus people with ASD can ameliorate colitis severity and associated sociability deficits.
- D. Garrett Brown
- Michaela Murphy
- June L. Round
Untargeted metabolomics-based network pharmacology reveals fermented brown rice towards anti-obesity efficacy
- Kaliyan Barathikannan
- Ramachandran Chelliah
- Deog- Hwan Oh
News and Comment

The ‘Mother Tree’ idea is everywhere — but how much of it is real?
A popular theory about how trees cooperate has enchanted the public and raised the profile of forest conservation. But some ecologists think its scientific basis has been oversold.
- Aisling Irwin
Bacteriophages and host inflammation in IBD
- Eleni Kotsiliti
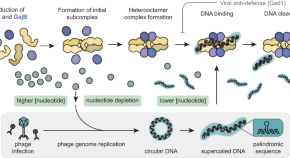
An OLD protein teaches us new tricks: prokaryotic antiviral defense
Reporting in Nature Communications , Huo and colleagues provide three-dimensional structures of a bacterial immune defense system called Gabija. This work builds on recently published structural and functional studies and contributes strong evidence that protein assembly formation is essential for antiviral function.
- Eirene Marie Q. Ednacot
- Benjamin R. Morehouse
Quick links
- Explore articles by subject
- Guide to authors
- Editorial policies
Microbiology News
Top headlines, latest headlines.
- Crimean-Congo Hemorrhagic Fever Virus
- Cancer Resistance in Tasmanian Devils
- Anti-Bacterial Medications to Treat TB
- Enzymatic Mix Kills TB-Causing Mycobacteria
- Silicon Spikes Take out 96% of Virus Particles
- Emerging Powassan Virus Spread by Deer Ticks
- Could Tardigrade Proteins Slow Aging in Humans?
- Maize Genes Control Little Helpers in the Soil
- Sensor Detects Dangerous Bacteria
- Treating Obstinate Mycobacterial Infections
Earlier Headlines
Monday, march 25, 2024.
- Humans Pass More Viruses to Other Animals Than We Catch from Them
- Researchers Discover Evolutionary 'tipping Point' In Fungi
Friday, March 22, 2024
- Natural Recycling at the Origin of Life
- Scientists Close in on TB Blood Test Which Could Detect Millions of Silent Spreaders
- Researchers Invent Artificial Intelligence Model to Design New Superbug-Fighting Antibiotics
Thursday, March 21, 2024
- As We Age, Our Cells Are Less Likely to Express Longer Genes
- Research Finds a Direct Communication Path Between the Lungs and the Brain
- Decoding the Plant World's Complex Biochemical Communication Networks
Wednesday, March 20, 2024
- Deep Earth Electrical Grid Mystery Solved
- Bacteria Subtype Linked to Growth in Up to 50% of Human Colorectal Cancers
- Experts Warn Climate Change Will Fuel Spread of Infectious Diseases
- Fiber, Genes and the Gut Microbiome: Study Reveals Possible Triggers for Inflammatory Bowel Disease
- Craving Snacks After a Meal? It Might Be Food-Seeking Neurons, Not an Overactive Appetite
Tuesday, March 19, 2024
- A Protein Found in Human Sweat May Protect Against Lyme Disease
Monday, March 18, 2024
- Climate Change Alters the Hidden Microbial Food Web in Peatlands
- Genes Identified That Allow Bacteria to Thrive Despite Toxic Heavy Metal in Soil
- Industrial Societies Losing Healthy Gut Microbes
- New Discovery Concerning Occurrence of Antibiotic Resistance
- Rise in Global Fungal Drug-Resistant Infections
- Cacao Plants' Defense Against Toxic Cadmium Unveiled
- Newborn Piglets Serve as a Model for Studying Influenza
- Engineers Measure pH in Cell Condensates
Friday, March 15, 2024
- 'Noisy' Roundworm Brains Give Rise to Individuality
- Gut Bacteria Make Neurotransmitters to Shape the Newborn Immune System
- Even Inactive Smokers Are Densely Colonized by Microbial Communities
- Protein Fragments ID Two New 'extremophile' Microbes--and May Help Find Alien Life
- Gut Bacteria Important for Overcoming Milk Allergy
Thursday, March 14, 2024
- Infections from These Bacteria Are on the Rise: New Blood Test Cuts Diagnosis Time from Months to Hours
- New Study on Mating Behaviors Offers Clues Into the Evolution of Attraction
- Dog-Killing Flatworm Discovered in Southern California
- It's Hearty, It's Meaty, It's Mold
- Alzheimer's Drug Fermented With Help from AI and Bacteria Moves Closer to Reality
- New Bioengineered Protein Design Shows Promise in Fighting COVID-19
- New Simpler and Cost-Effective Forensics Test Helps Identify Touch DNA
Wednesday, March 13, 2024
- Sulfur and the Origin of Life
- Tryptophan in Diet, Gut Bacteria Protect Against E. Coli Infection
- Study Shows Important Role Gut Microbes Play in Airway Health in Persons With Cystic Fibrosis
- Steroid Drugs Used for HRT Can Combat E. Coli and MRSA
- Simple Trick Could Improve Accuracy of Plant Genetics Research
- Milk to the Rescue for Diabetics? Cow Produces Human Insulin in Milk
- Who Knew That Coprophagy Was So Vital for Birds' Survival?
Tuesday, March 12, 2024
- A Simple and Robust Experimental Process for Protein Engineering
- Scientists Find Weak Points on Epstein-Barr Virus
- Maternal Obesity May Promote Liver Cancer
Monday, March 11, 2024
- Researchers Uncover Protein Responsible for Cold Sensation
- Higher Bacterial Counts Detected in Single-Serving Milks, Researchers Report
Friday, March 8, 2024
- New Study Discovers How Altered Protein Folding Drives Multicellular Evolution
- Researchers Develop Artificial Building Blocks of Life
- Researchers Open New Leads in Anti-HIV Drug Development, Using a Compound Found in Nature
Thursday, March 7, 2024
- How Does a Virus Hijack Insect Sperm to Control Disease Vectors and Pests?
- The Malaria Parasite Generates Genetic Diversity Using an Evolutionary 'copy-Paste' Tactic
- What Makes a Pathogen Antibiotic-Resistant?
- First Atom-Level Structure of Packaged Viral Genome Reveals New Properties, Dynamics
- Cracking Epigenetic Inheritance: Biologists Discovered the Secrets of How Gene Traits Are Passed on
Wednesday, March 6, 2024
- Revealing a Hidden Threat: Researchers Show Viral Infections Pose Early Heart Risks
- Microbes Impact Coral Bleaching Susceptibility
- Early Life Adversity Leaves Long-Term Signatures in Baboon DNA
- Revealing the Evolutionary Origin of Genomic Imprinting
- Universal Tool for Tracking Cell-to-Cell Interactions
- Synthetic Gene Helps Explain the Mysteries of Transcription Across Species
- Decoding the Language of Epigenetic Modifications
- Deconstructing the Structural Elements of a Lesser-Known Microbe
Tuesday, March 5, 2024
- Lab-Grown Liver Organoid to Speed Up Turtle Research, Making Useful Traits Easier to Harness
- Possible 'Trojan Horse' Found for Treating Stubborn Bacterial Infections
- Microalgae With Unusual Cell Biology
Monday, March 4, 2024
- Protecting Joints from Bacteria With Mussels
- Modeling the Origins of Life: New Evidence for an 'RNA World'
- An Evolutionary Mystery 125 Million Years in the Making
- Photosynthetic Secrets Come to Light
- Advances in Forensic Science Improve Accuracy of 'time of Death' Estimates
Friday, March 1, 2024
- Researchers Create Coating Solution for Safer Food Storage
- Light Into the Darkness of Photosynthesis
- How Virus Causes Cancer: Potential Treatment
Thursday, February 29, 2024
- New Role for Bacterial Enzyme in Gut Metabolism Revealed
- Refrigerate Lettuce to Reduce Risk of E. Coli Contamination
- Microbial Viruses Act as Secret Drivers of Climate Change
- Measuring Electrical Conductivity in Microorganisms, Approaching Understanding of Microbial Ecosystems
- Scientists Develop Novel RNA Or DNA-Based Substances to Protect Plants from Viruses
- Radio Waves Can Tune Up Bacteria to Become Life-Saving Medicines
- Scientists Discover 18 New Species of Gut Microbes in Search for Origins of Antibiotic Resistance
Wednesday, February 28, 2024
- Study Reveals Accelerated Soil Priming Under Climate Warming
- New Tool Helps Decipher Gene Behavior
- In Fight Against Brain Pathogens, the Eyes Have It
- Double Trouble at Chromosome Ends
- Change in Gene Code May Explain How Human Ancestors Lost Tails
Tuesday, February 27, 2024
- New Discovery Shows How Cells Defend Themselves During Stressful Situations
- Microbial Comics: RNA as a Common Language, Presented in Extracellular Speech-Bubbles
- Low-Temperature Plasma Used to Remove E. Coli from Hydroponically Grown Crops
Monday, February 26, 2024
- 'Janitors' Of the Sea: Overharvested Sea Cucumbers Play Crucial Role in Protecting Coral
- Cutting-Edge 'protein Lawnmower' Created
- Scientists Assemble a Richer Picture of the Plight and Resilience of the Foothill Yellow-Legged Frog
- Blindness from Some Inherited Eye Diseases May Be Caused by Gut Bacteria
Sunday, February 25, 2024
- Global Warming Increases the Diversity of Active Soil Bacteria
Friday, February 23, 2024
- Altering the Circadian Clock Adapts Barley to Short Growing Seasons
- Lab-Spun Sponges Form Perfect Scaffolds for Growing Skin Cells to Heal Wounds
Thursday, February 22, 2024
- 'Dynamic Duo' Defenses in Bacteria Ward Off Viral Threats
- Compound Vital for All Life Likely Played a Role in Life's Origin
- Copies of Antibiotic Resistance Genes Greatly Elevated in Humans and Livestock
- Metabolic Diseases May Be Driven by Gut Microbiome, Loss of Ovarian Hormones
- Damage to Cell Membranes Causes Cell Aging
- LATEST NEWS
- Top Science
- Top Physical/Tech
- Top Environment
- Top Society/Education
- Health & Medicine
- Mind & Brain
- Living Well
- Space & Time
- Matter & Energy
- Computers & Math
- Plants & Animals
- Agriculture & Food
- Beer and Wine
- Bird Flu Research
- Genetically Modified
- Pests and Parasites
- Cows, Sheep, Pigs
- Dolphins and Whales
- Frogs and Reptiles
- Insects (including Butterflies)
- New Species
- Spiders and Ticks
- Veterinary Medicine
- Business & Industry
- Biotechnology and Bioengineering
- CRISPR Gene Editing
- Food and Agriculture
- Endangered Animals
- Endangered Plants
- Extreme Survival
- Invasive Species
- Wild Animals
- Education & Learning
- Animal Learning and Intelligence
- Life Sciences
- Behavioral Science
- Biochemistry Research
- Biotechnology
- Cell Biology
- Developmental Biology
- Epigenetics Research
- Evolutionary Biology
- Marine Biology
- Mating and Breeding
- Molecular Biology
- Microbes and More
- Microbiology
- Zika Virus Research
- Earth & Climate
- Fossils & Ruins
- Science & Society
Strange & Offbeat
- Illuminating Oxygen's Journey in the Brain
- DNA Study IDs Descendants of George Washington
- Heart Disease Risk: More Than One Drink a Day
- Unlocking Supernova Stardust Secrets
- Why Do Some Memories Become Longterm?
- Cell Division Quality Control 'Stopwatch'
- What Controls Sun's Differential Rotation?
- Robot, Can You Say 'Cheese'?
- Researchers Turn Back the Clock On Cancer Cells
- Making Long-Term Memories: Nerve-Cell Damage
Trending Topics
- Search by keyword
- Search by citation
Page 1 of 109
Bacillus subtilis SOM8 isolated from sesame oil meal for potential probiotic application in inhibiting human enteropathogens
While particular strains within the Bacillus species, such as Bacillus subtilis , have been commercially utilised as probiotics, it is critical to implement screening assays and evaluate the safety to identify pot...
- View Full Text
Promiscuous, persistent and problematic: insights into current enterococcal genomics to guide therapeutic strategy
Vancomycin-resistant enterococci (VRE) are major opportunistic pathogens and the causative agents of serious diseases, such as urinary tract infections and endocarditis. VRE strains mainly include species of Ente...
Comparison of integron mediated antimicrobial resistance in clinical isolates of Escherichia coli from urinary and bacteremic sources
Antimicrobial resistance (AMR) is a global threat driven mainly by horizontal gene transfer (HGT) mechanisms through mobile genetic elements (MGEs) including integrons. The variable region (VR) of an integron ...
Structure predictions and functional insights into Amidase_3 domain containing N -acetylmuramyl-L-alanine amidases from Deinococcus indicus DR1
N -acetylmuramyl-L-alanine amidases are cell wall modifying enzymes that cleave the amide bond between the sugar residues and stem peptide in peptidoglycan. Amidases play a vital role in septal cell wall cleavag.....
Profile of non-tuberculous mycobacteria amongst tuberculosis presumptive people in Cameroon
Cameroon is a tuberculosis (TB) burden country with a 12% positivity among TB presumptive cases. Of the presumptive cases with a negative TB test, some are infected with Non-tuberculous Mycobacteria (NTM). How...
In vitro investigation of relationship between quorum-sensing system genes, biofilm forming ability, and drug resistance in clinical isolates of Pseudomonas aeruginosa
Pseudomonas aeruginosa is an opportunistic pathogen in the health-care systems and one of the primary causative agents with high mortality in hospitalized patients, particularly immunocompromised. The limitation ...
Relationship between heart failure and intestinal inflammation in infants with congenital heart disease
The association between heart failure (HF) and intestinal inflammation caused by a disturbed intestinal microbiota in infants with congenital heart disease (CHD) was investigated.
Clostridium butyricum inhibits the inflammation in children with primary nephrotic syndrome by regulating Th17/Tregs balance via gut-kidney axis
Primary nephrotic syndrome (PNS) is a common glomerular disease in children. Clostridium butyricum ( C. butyricum), a probiotic producing butyric acid, exerts effective in regulating inflammation. This study was d...
Human-derived bacterial strains mitigate colitis via modulating gut microbiota and repairing intestinal barrier function in mice
Unbalanced gut microbiota is considered as a pivotal etiological factor in colitis. Nevertheless, the precise influence of the endogenous gut microbiota composition on the therapeutic efficacy of probiotics in...
In vitro and in silico studies of enterobactin-inspired Ciprofloxacin and Fosfomycin first generation conjugates on the antibiotic resistant E. coli OQ866153
The emergence of antimicrobial resistance in bacterial pathogens is a growing concern worldwide due to its impact on the treatment of bacterial infections. The "Trojan Horse" strategy has been proposed as a po...
HPV-associated cervicovaginal microbiome and host metabolome characteristics
Cervicovaginal microbiome plays an important role in the persistence of HPV infection and subsequent disease development. However, cervicovaginal microbiota varied cross populations with different habits and r...
Transcriptomic and physiological analyses of Trichoderma citrinoviride HT-1 assisted phytoremediation of Cd contaminated water by Phragmites australis
Plant growth promoting microbe assisted phytoremediation is considered a more effective approach to rehabilitation than the single use of plants, but underlying mechanism is still unclear. In this study, we co...
Long-term push–pull cropping system shifts soil and maize-root microbiome diversity paving way to resilient farming system
The soil biota consists of a complex assembly of microbial communities and other organisms that vary significantly across farming systems, impacting soil health and plant productivity. Despite its importance, ...
Pretreatment with an antibiotics cocktail enhances the protective effect of probiotics by regulating SCFA metabolism and Th1/Th2/Th17 cell immune responses
Probiotics are a potentially effective therapy for inflammatory bowel disease (IBD); IBD is linked to impaired gut microbiota and intestinal immunity. However, the utilization of an antibiotic cocktail (Abx) p...
High-throughput sequencing reveals differences in microbial community structure and diversity in the conjunctival tissue of healthy and type 2 diabetic mice
To investigate the differences in bacterial and fungal community structure and diversity in conjunctival tissue of healthy and diabetic mice.
High prevalence of ST5-SCC mec II-t311 clone of methicillin-resistant Staphylococcus aureus isolated from bloodstream infections in East China
Methicillin-resistant Staphylococcus aureus (MRSA) is a challenging global health threat, resulting in significant morbidity and mortality worldwide. This study aims to determine the molecular characteristics and...
Characteristics of the oral and gastric microbiome in patients with early-stage intramucosal esophageal squamous cell carcinoma
Oral microbiome dysbacteriosis has been reported to be associated with the pathogenesis of advanced esophageal cancer. However, few studies investigated the potential role of oral and gastric microbiota in ear...
The potential role of Listeria monocytogenes in promoting colorectal adenocarcinoma tumorigenic process
Listeria monocytogenes is a foodborne pathogen, which can cause a severe illness, especially in people with a weakened immune system or comorbidities. The interactions between host and pathogens and between patho...
Evaluation of clinical characteristics and risk factors associated with Chlamydia psittaci infection based on metagenomic next-generation sequencing
Psittacosis is a zoonosis caused by Chlamydia psittaci , the clinical manifestations of Psittacosis range from mild illness to fulminant severe pneumonia with multiple organ failure. This study aimed to evaluate t...
Characterization of the broad-spectrum antibacterial activity of bacteriocin-like inhibitory substance-producing probiotics isolated from fermented foods
Antimicrobial peptides, such as bacteriocin, produced by probiotics have become a promising novel class of therapeutic agents for treating infectious diseases. Selected lactic acid bacteria (LAB) isolated from...
Metagenomic gut microbiome analysis of Japanese patients with multiple chemical sensitivity/idiopathic environmental intolerance
Although the pathology of multiple chemical sensitivity (MCS) is unknown, the central nervous system is reportedly involved. The gut microbiota is important in modifying central nervous system diseases. Howeve...
The effect of in vitro simulated colonic pH gradients on microbial activity and metabolite production using common prebiotics as substrates
The interplay between gut microbiota (GM) and the metabolization of dietary components leading to the production of short-chain fatty acids (SCFAs) is affected by a range of factors including colonic pH and ca...
Gut microbial network signatures of early colonizers in preterm neonates with extrauterine growth restriction
Extrauterine growth restriction (EUGR) represents a prevalent condition observed in preterm neonates, which poses potential adverse implications for both neonatal development and long-term health outcomes. The...
Early transcriptional changes of heavy metal resistance and multiple efflux genes in Xanthomonas campestris pv. campestris under copper and heavy metal ion stress
Copper-induced gene expression in Xanthomonas campestris pv. campestris (Xcc) is typically evaluated using targeted approaches involving qPCR. The global response to copper stress in Xcc and resistance to metal i...
Whole-genome sequencing and analysis of Chryseobacterium arthrosphaerae from Rana nigromaculata
Chryseobacterium arthrosphaerae strain FS91703 was isolated from Rana nigromaculata in our previous study. To investigate the genomic characteristics, pathogenicity-related genes, antimicrobial resistance, and ph...
Changes in the nasopharyngeal and oropharyngeal microbiota in pediatric obstructive sleep apnea before and after surgery: a prospective study
To explore the changes and potential mechanisms of microbiome in different parts of the upper airway in the development of pediatric OSA and observe the impact of surgical intervention on oral microbiome for p...
Unveiling biological activities of biosynthesized starch/silver-selenium nanocomposite using Cladosporium cladosporioides CBS 174.62
Microbial cells capability to tolerate the effect of various antimicrobial classes represent a major worldwide health concern. The flexible and multi-components nanocomposites have enhanced physicochemical cha...
Characterization of the major autolysin ( AtlC ) of Staphylococcus carnosus
Autolysis by cellular peptidoglycan hydrolases (PGH) is a well-known phenomenon in bacteria. During food fermentation, autolysis of starter cultures can exert an accelerating effect, as described in many studi...
Exploring Aeromonas dhakensis in Aldabra giant tortoises: a debut report and genetic characterization
Aeromonas dhakensis (A. dhakensis) is becoming an emerging pathogen worldwide, with an increasingly significant role in animals and human health. It is a ubiquitous bacteria found in terrestrial and aquatic milie...
Biotransformation of zearalenone to non-estrogenic compounds with two novel recombinant lactonases from Gliocladium
The mycotoxin zearalenone (ZEA) produced by toxigenic fungi is widely present in cereals and its downstream products. The danger of ZEA linked to various human health issues has attracted increasing attention....
Multidrug resistance among uropathogenic clonal group A E. Coli isolates from Pakistani women with uncomplicated urinary tract infections
Multi-drug resistance (MDR) has notably increased in community acquired uropathogens causing urinary tract infections (UTIs), predominantly Escherichia coli . Uropathogenic E. coli causes 80% of uncomplicated comm...
Uncovering the complexity of childhood undernutrition through strain-level analysis of the gut microbiome
Undernutrition (UN) is a critical public health issue that threatens the lives of children under five in developing countries. While evidence indicates the crucial role of the gut microbiome (GM) in UN pathoge...
RpoS role in antibiotic resistance, tolerance and persistence in E. coli natural isolates
The intrinsic concentration of RpoS, the second most abundant sigma factor, varies widely across the E. coli species. Bacterial isolates that express high levels of RpoS display high resistance to environmental s...
Fine-scale geographic difference of the endangered Big-headed Turtle ( Platysternon megacephalum ) fecal microbiota, and comparison with the syntopic Beale’s Eyed Turtle ( Sacalia bealei )
Studies have elucidated the importance of gut microbiota for an organism, but we are still learning about the important influencing factors. Several factors have been identified in helping shape the microbiome...
The activation impact of lactobacillus-derived extracellular vesicles on lipopolysaccharide-induced microglial cell
Perioperative neurocognitive dysfunction (PND) emerges as a common postoperative complication among elderly patients. Currently, the mechanism of PND remains unclear, but there exists a tendency to believe tha...
Development of non-alcoholic steatohepatitis is associated with gut microbiota but not with oxysterol enzymes CH25H, EBI2, or CYP7B1 in mice
Liver steatosis is the most frequent liver disorder and its advanced stage, non-alcoholic steatohepatitis (NASH), will soon become the main reason for liver fibrosis and cirrhosis. The “multiple hits hypothesi...
Rapid, visual, label-based biosensor platform for identification of hepatitis C virus in clinical applications
In the current study, for the first time, we reported a novel HCV molecular diagnostic approach termed reverse transcription loop-mediated isothermal amplification integrated with a gold nanoparticles-based la...
Phenotypic, molecular, and in silico characterization of coumarin as carbapenemase inhibitor to fight carbapenem-resistant Klebsiella pneumoniae
Carbapenems represent the first line treatment of serious infections caused by drug-resistant Klebsiella pneumoniae . Carbapenem-resistant K. pneumoniae (CRKP) is one of the urgent threats to human health worldwid...
Transcriptomic meta-analysis to identify potential antifungal targets in Candida albicans
Candida albicans is a fungal pathogen causing human infections. Here we investigated differential gene expression patterns and functional enrichment in C. albicans strains grown under different conditions.
Escherichia coli and their potential transmission of carbapenem and colistin-resistant genes in camels
Camels harbouring multidrug-resistant Gram-negative bacteria are capable of transmitting various microorganisms to humans. This study aimed to determine the distribution of anti-microbial resistance among Escheri...
Difference analysis and characteristics of incompatibility group plasmid replicons in gram-negative bacteria with different antimicrobial phenotypes in Henan, China
Multi-drug-resistant organisms (MDROs) in gram-negative bacteria have caused a global epidemic, especially the bacterial resistance to carbapenem agents. Plasmid is the common vehicle for carrying antimicrobia...
Development of a novel mycobiome diagnostic for fungal infection
Amplicon-based mycobiome analysis has the potential to identify all fungal species within a sample and hence could provide a valuable diagnostic assay for use in clinical mycology settings. In the last decade,...
Effects of the alpine meadow in different phenological periods on rumen fermentation and gastrointestinal tract bacteria community in grazing yak on the Qinghai-Tibetan Plateau
In this study, we investigated the effects of alpine meadow in different phenological periods on ruminal fermentation, serum biochemical indices, and gastrointestinal tract microbes in grazing yak on the Qingh...
Emergence of multidrug-resistant Bacillus spp. derived from animal feed, food and human diarrhea in South-Eastern Bangladesh
Antimicrobial resistance poses a huge risk to human health worldwide, while Bangladesh is confronting the most severe challenge between the food supply and the huge consumption of antibiotics annually. More im...
What happens to Bifidobacterium adolescentis and Bifidobacterium longum ssp. longum in an experimental environment with eukaryotic cells?
The impact of probiotic strains on host health is widely known. The available studies on the interaction between bacteria and the host are focused on the changes induced by bacteria in the host mainly. The stu...
Outbreak of colistin and carbapenem-resistant Klebsiella pneumoniae ST16 co-producing NDM-1 and OXA-48 isolates in an Iranian hospital
Colistin and carbapenem-resistant Klebsiella pneumoniae (Col-CRKP) represent a significant and constantly growing threat to global public health. We report here an outbreak of Col-CRKP infections during the fifth...
Optimized bacterial community characterization through full-length 16S rRNA gene sequencing utilizing MinION nanopore technology
Accurate identification of bacterial communities is crucial for research applications, diagnostics, and clinical interventions. Although 16S ribosomal RNA (rRNA) gene sequencing is a widely employed technique ...
Metagenomic analysis of gut microbiome illuminates the mechanisms and evolution of lignocellulose degradation in mangrove herbivorous crabs
Sesarmid crabs dominate mangrove habitats as the major primary consumers, which facilitates the trophic link and nutrient recycling in the ecosystem. Therefore, the adaptations and mechanisms of sesarmid crabs...

ST218 Klebsiella pneumoniae became a high-risk clone for multidrug resistance and hypervirulence
The occurrence of multidrug-resistant and hypervirulent Klebsiella pneumoniae (MDR-hvKp) worldwide poses a great challenge for public health. Few studies have focused on ST218 MDR-hvKp.
The anti-staphylococcal fusidic acid as an efflux pump inhibitor combined with fluconazole against vaginal candidiasis in mouse model
Candida albicans is the most common fungus that causes vaginal candidiasis in immunocompetent women and catastrophic infections in immunocompromised patients. The treatment of such infections is hindered due to t...
Important information
Editorial board
For authors
For editorial board members
For reviewers
- Manuscript editing services
Annual Journal Metrics
2022 Citation Impact 4.2 - 2-year Impact Factor 4.7 - 5-year Impact Factor 1.131 - SNIP (Source Normalized Impact per Paper) 0.937 - SJR (SCImago Journal Rank)
2023 Speed 19 days submission to first editorial decision for all manuscripts (Median) 135 days submission to accept (Median)
2023 Usage 2,970,572 downloads 1,619 Altmetric mentions
- More about our metrics
- Follow us on Twitter
BMC Microbiology
ISSN: 1471-2180
- General enquiries: [email protected]
Microbiology
Viral Genetics Confirms What On-the-Ground Activists Knew Early in the Mpox Outbreak
Molecular biology could have changed the mpox epidemic—and could stop future outbreaks
Joseph Osmundson
Cannibal Cells Inspire Cancer Treatment Improvement
Giving cells an appetite for cancer could enhance treatments
Kate Graham-Shaw

Is Raw-Milk Cheese Safe to Eat?
Recent bacterial outbreaks from consuming cheese made from unpasteurized milk, or “raw milk,” raise questions about the safety of eating these artisanal products
Riis Williams
Many Pregnancy Losses Are Caused by Errors in Cell Division
Odd cell divisions could help explain why even young, healthy couples might struggle to get pregnant
Gina Jiménez

'Microbiome of Death' Uncovered on Decomposing Corpses Could Aid Forensics
Microbes that lurk in decomposing human corpses could help forensic detectives establish a person's time of death
Christoph Schwaiger, LiveScience
Weird ‘Obelisks’ Found in Human Gut May be Virus-Like Entities
Rod-shaped fragments of RNA called “obelisks” were discovered in gut and mouth bacteria for the first time
Joanna Thompson
Semen Has Its Own Microbiome—And It Might Influence Fertility
Recent research found a species of bacteria living in semen that’s associated with infertility and has links to the vaginal microbiome
Andrew Chapman

Bacteria Make Decisions Based on Generational Memories
Bacteria choose to swarm based on what happened to their great-grandparents
Allison Parshall

Your Body Has Its Own Built-In Ozempic
Popular weight-loss and diabetes drugs, such as Ozempic and Wegovy, target metabolic pathways that gut microbes and food molecules already play a key role in regulating
Christopher Damman, The Conversation US

See Your Body’s Cells in Size and Number
The larger a cell type is, the rarer it is in the body—and vice versa—a new study shows
Clara Moskowitz, Jen Christiansen, Ni-ka Ford

Subterranean ‘Microbial Dark Matter’ Reveals a Strange Dichotomy
The genes of microbes living as deep as 1.5 kilometers below the surface reveal a split between minimalist and maximalist lifestyles
Stephanie Pappas
The Vaginal Microbiome May Affect Health More than We Thought
A recent study finds varying combinations of microbes in the vaginal microbiome may influence health outcomes such as risk of sexually transmitted disease and preterm birth
Lori Youmshajekian
Suggestions or feedback?
MIT News | Massachusetts Institute of Technology
- Machine learning
- Social justice
- Black holes
- Classes and programs
Departments
- Aeronautics and Astronautics
- Brain and Cognitive Sciences
- Architecture
- Political Science
- Mechanical Engineering
Centers, Labs, & Programs
- Abdul Latif Jameel Poverty Action Lab (J-PAL)
- Picower Institute for Learning and Memory
- Lincoln Laboratory
- School of Architecture + Planning
- School of Engineering
- School of Humanities, Arts, and Social Sciences
- Sloan School of Management
- School of Science
- MIT Schwarzman College of Computing
Turning microbiome research into a force for health
Press contact :.
Previous image Next image
The microbiome comprises trillions of microorganisms living on and inside each of us. Historically, researchers have only guessed at its role in human health, but in the last decade or so, genetic sequencing techniques have illuminated this galaxy of microorganisms enough to study in detail.
As researchers unravel the complex interplay between our bodies and microbiomes, they are beginning to appreciate the full scope of the field’s potential for treating disease and promoting health.
For instance, the growing list of conditions that correspond with changes in the microbes of our gut includes type 2 diabetes, inflammatory bowel disease, Alzheimer’s disease, and a variety of cancers.
“In almost every disease context that’s been investigated, we’ve found different types of microbial communities, divergent between healthy and sick patients,” says professor of biological engineering Eric Alm. “The promise [of these findings] is that some of those differences are going to be causal, and intervening to change the microbiome is going to help treat some of these diseases.”
Alm’s lab, in conjunction with collaborators at the Broad Institute of MIT and Harvard, did some of the early work characterizing the gut microbiome and showing its relationship to human health. Since then, microbiome research has exploded, pulling in researchers from far-flung fields and setting new discoveries in motion. Startups are now working to develop microbiome-based therapies, and nonprofit organizations have also sprouted up to ensure these basic scientific advances turn into treatments that benefit the maximum number of people.
“The first chapter in this field, and our history, has been validating this modality,” says Mark Smith PhD ’14, a co-founder of OpenBiome, which processes stool donations for hospitals to conduct stool transplants for patients battling gut infection. Smith is also currently CEO of the startup Finch Therapeutics, which is developing microbiome-based treatments. “Until now, it’s been about the promise of the microbiome. Now I feel like we’ve delivered on the first promise. The next step is figuring out how big this gets.”
An interdisciplinary foundation
MIT’s prominent role in microbiome research came, in part, through its leadership in a field that may at first seem unrelated. For decades, MIT has made important contributions to microbial ecology, led by work in the Parsons Laboratory in the Department of Civil and Environmental Engineering and by scientists including Institute Professor Penny Chisholm.
Ecologists who use complex statistical techniques to study the relationships between organisms in different ecosystems are well-equipped to study the behavior of different bacterial strains in the microbiome.
Not that ecologists — or anyone else — initially had much to study involving the human microbiome, which was essentially a black box to researchers well into the 2000s. But the Human Genome Project led to faster, cheaper ways to sequence genes at scale, and a group of researchers including Alm and visiting professor Martin Polz began using those techniques to decode the genomes of environmental bacteria around 2008.
Those techniques were first pointed at the bacteria in the gut microbiome as part of the Human Microbiome Project, which began in 2007 and involved research groups from MIT and the Broad Institute.
Alm first got pulled into microbiome research by the late biological engineering professor David Schauer as part of a research project with Boston Children’s Hospital. It didn’t take much to get up to speed: Alm says the number of papers explicitly referencing the microbiome at the time could be read in an afternoon.
The collaboration, which included Ramnik Xavier, a core institute member of the Broad Institute, led to the first large-scale genome sequencing of the gut microbiome to diagnose inflammatory bowel disease. The research was funded, in part, by the Neil and Anna Rasmussen Family Foundation.
The study offered a glimpse into the microbiome’s diagnostic potential. It also underscored the need to bring together researchers from diverse fields to dig deeper.
Taking an interdisciplinary approach is important because, after next-generation sequencing techniques are applied to the microbiome, a large amount of computational biology and statistical methods are still needed to interpret the resulting data — the microbiome, after all, contains more genes than the human genome. One catalyst for early microbiome collaboration was the Microbiology Graduate PhD Program, which recruited microbiology students to MIT and introduced them to research groups across the Institute.
As microbiology collaborations increased among researchers from different department and labs, Neil Rasmussen, a longtime member of the MIT Corporation and a member of the visiting committees for a number of departments, realized there was still one more component needed to turn microbiome research into a force for human health.
“Neil had the idea to find all the clinical researchers in the [Boston] area studying diseases associated with the microbiome and pair them up with people like [biological engineers, mathematicians, and ecologists] at MIT who might not know anything about inflammatory bowel disease or microbiomes but had the expertise necessary to solve big problems in the field,” Alm says.
In 2014, that insight led the Rasmussen Foundation to support the creation of the Center for Microbiome Informatics and Therapeutics (CMIT), one of the first university-based microbiome research centers in the country. CMIT is based at the MIT Institute for Medical Engineering and Science (IMES).
Tami Lieberman, the Hermann L. F. von Helmholtz Career Development Professor at MIT, whose background is in ecology, says CMIT was a big reason she joined MIT’s faculty in 2018. Lieberman has developed new genomic approaches to study how bacteria mutate in healthy and sick individuals, with a particular focus on the skin microbiome.
Laura Kiessling, a chemist who has been recognized for contributions to our understanding of cell surface interactions, was also quick to join CMIT. Kiessling, the Novartis Professor of Chemistry, has made discoveries relating to microbial mechanisms that influence immune function. Both Lieberman and Kiessling are also members of the Broad Institute.
Today, CMIT, co-directed by Alm and Xavier, facilitates collaborations between researchers and clinicians from hospitals around the country in addition to supporting research groups in the area. That work has led to hundreds of ongoing clinical trials that promise to further elucidate the microbiome’s connection to a broad range of diseases.
Fulfilling the promise of the microbiome
Researchers don’t yet know what specific strains of bacteria can improve the health of people with microbiome-associated diseases. But they do know that fecal matter transplants, which carry the full spectrum of gut bacteria from a healthy donor, can help patients suffering from certain diseases.
The nonprofit organization OpenBiome, founded by a group from MIT including Smith and Alm, launched in 2012 to help expand access to fecal matter transplants by screening donors for stool collection then processing, storing, and shipping samples to hospitals. Today OpenBiome works with more than 1,000 hospitals, and its success in the early days of the field shows that basic microbiome research, when paired with clinical trials like those happening at CMIT, can quickly lead to new treatments.
“You start with a disease, and if there’s a microbiome association, you can start a small trial to see if fecal transplants can help patients right away,” Alm explains. “If that becomes an effective treatment, while you’re rolling it out you can be doing the genomics to figure out how to make it better. So you can translate therapeutics into patients more quickly than when you’re developing small-molecule drugs.”
Another nonprofit project launched out of MIT, the Global Microbiome Conservancy, is collecting stool samples from people living nonindustrialized lifestyles around the world, whose guts have much different bacterial makeups and thus hold potential for advancing our understanding of host-microbiome interactions.
A number of private companies founded by MIT alumni are also trying to harness individual microbes to create new treatments, including, among others, Finch Therapeutics founded by Mark Smith; Concerto Biosciences, co-founded by Jared Kehe PhD ’20 and Bernardo Cervantes PhD ’20; BiomX, founded by Associate Professor Tim Lu; and Synlogic, founded by Lu and Jim Collins, the Termeer Professor of Medical Engineering and Science at MIT.
“There’s an opportunity to more precisely change a microbiome,” explains CMIT’s Lieberman. “But there’s a lot of basic science to do to figure out how to tweak the microbiome in a targeted way. Once we figure out how to do that, the therapeutic potential of the microbiome is quite limitless.”
Share this news article on:
Related links.
- MIT Center for Microbiome Informatics and Therpeutics
- Lieberman Lab
- Broad Institute
- Department of Biological Engineering
- Department of Civil and Environmental Engineering
Related Topics
- Biological engineering
- Innovation and Entrepreneurship (I&E)
- Drug development
- Institute for Medical Engineering and Science (IMES)
Related Articles
![recent research on microbiology “The biology [around gut bacteria’s influence on health] is fairly complex, and we’re still in the early days of unravelling it, but there have been a number of clinical studies that have reported benefits to restoring gut health, and that’s our north star: the clinical data,” Finch co-founder and Chief Executive Officer Mark Smith PhD ’14 says.](https://news.mit.edu/sites/default/files/styles/news_article__archive/public/images/202007/MIT-Finch-01.jpg?itok=nMuVS-Q9)
Finch Therapeutics unleashes the power of the gut

A comprehensive catalogue of human digestive tract bacteria

3Q: Eric Alm on the mysteries of the microbiome

Studying the hotbed of horizontal gene transfers

MIT startup tackles nasty infection with first public stool bank
Previous item Next item
More MIT News
A first-ever complete map for elastic strain engineering
Read full story →

“Life is short, so aim high”

Shining a light on oil fields to make them more sustainable

MIT launches Working Group on Generative AI and the Work of the Future

Atmospheric observations in China show rise in emissions of a potent greenhouse gas

Second round of seed grants awarded to MIT scholars studying the impact and applications of generative AI
- More news on MIT News homepage →
Massachusetts Institute of Technology 77 Massachusetts Avenue, Cambridge, MA, USA
- Map (opens in new window)
- Events (opens in new window)
- People (opens in new window)
- Careers (opens in new window)
- Accessibility
- Social Media Hub
- MIT on Facebook
- MIT on YouTube
- MIT on Instagram
An official website of the United States government
The .gov means it’s official. Federal government websites often end in .gov or .mil. Before sharing sensitive information, make sure you’re on a federal government site.
The site is secure. The https:// ensures that you are connecting to the official website and that any information you provide is encrypted and transmitted securely.
- Publications
- Account settings
Preview improvements coming to the PMC website in October 2024. Learn More or Try it out now .
- Advanced Search
- Journal List
- Front Microbiol
The gut microbiome in human health and disease—Where are we and where are we going? A bibliometric analysis
Associated data.
The original contributions presented in the study are included in the article/ Supplementary material , further inquiries can be directed to the corresponding author.
There are trillions of microbiota in our intestinal tract, and they play a significant role in health and disease via interacting with the host in metabolic, immune, neural, and endocrine pathways. Over the past decades, numerous studies have been published in the field of gut microbiome and disease. Although there are narrative reviews of gut microbiome and certain diseases, the whole field is lack of systematic and quantitative analysis. Therefore, we outline research status of the gut microbiome and disease, and present insights into developments and characteristics of this field to provide a holistic grasp and future research directions.
An advanced search was carried out in the Web of Science Core Collection (WoSCC), basing on the term “gut microbiome” and its synonyms. The current status and developing trends of this scientific domain were evaluated by bibliometric methodology. CiteSpace was used to perform collaboration network analysis, co-citation analysis and citation burst detection.
A total of 29,870 articles and 13,311 reviews were retrieved from the database, which involve 42,900 keywords, 176 countries/regions, 19,065 institutions, 147,225 authors and 4,251 journals. The gut microbiome and disease research is active and has received increasing attention. Co-cited reference analysis revealed the landmark articles in the field. The United States had the largest number of publications and close cooperation with other countries. The current research mainly focuses on gastrointestinal diseases, such as inflammatory bowel disease (IBD), ulcerative colitis (UC) and Crohn’s disease (CD), while extra-intestinal diseases are also rising, such as obesity, diabetes, cardiovascular disease, Alzheimer’s disease, Parkinson’s disease. Omics technologies, fecal microbiota transplantation (FMT) and metabolites linked to mechanism would be more concerned in the future.
The gut microbiome and disease has been a booming field of research, and the trend is expected to continue. Overall, this research field shows a multitude of challenges and great opportunities.
Introduction
The human gut microbiota originated from colonization by environmental microbes during birth, and live in symbiosis with the host throughout life ( Koenig et al., 2011 ; Kundu et al., 2017 ). The inoculum source usually and mainly is the mother’s vaginal and fecal microbiomes ( Koenig et al., 2011 ). Human microbiota carried diverse set of genomes, and is considered as human second genome ( Grice and Segre, 2012 ). While the microbes that reside in our gut account for the vast majority, present more than 1,000 species ( Almeida et al., 2019 ), and the number of microorganisms is estimated up to trillions ( Sender et al., 2016 ). These abundant and diverse gut microbes constitute a dynamic and complex ecosystem and perform various functions that are essential for the human host ( Heintz-Buschart and Wilmes, 2018 ). On the one hand, there are competition and cooperation within these microbial consortia ( Coyte and Rakoff-Nahoum, 2019 ), on the other hand, they also interact with the host in multiple aspects, including digestion and metabolism ( Krautkramer et al., 2021 ), immune system ( Rooks and Garrett, 2016 ) and unconscious system ( Dinan and Cryan, 2017 ). Hence, the gut microbiome directly or indirectly impacts the host’s health.
It should be noted that the concept that our resident microbial communities make essential contributions to the host’s physiology and health can date back to Louis Pasteur (1822–1895; Stappenbeck et al., 2002 ). Indeed, the gut microbiome has been associated with various diseases and conditions in the past decades, such as IBD ( Morgan et al., 2012 ), obesity ( Fei and Zhao, 2013 ), diabetes ( Lau et al., 2021 ), Parkinson’s disease ( Wallen et al., 2021 ) and cancer ( Gopalakrishnan et al., 2018 ). Meanwhile, the gut microbiome shows great promise for disease diagnosis, i.e., as microbial biomarkers with operational taxonomic units (OTUs), taxa and metabolite ( Wu et al., 2021 ); and for disease therapy by manipulation of the gut microbiome, such as dietary interventions, microbial supplements and FMT ( Durack and Lynch, 2018 ).
The role of the gut microbiome in human health and disease has received increasing attention over the last 20 years, and the trend is expected to continue. At present, some fundamental problems need to be addressed in this field. For example, the taxa, genome, functions and cultivation of microbial dark matter ( Pasolli et al., 2019 ; Jiao et al., 2021 ). Moreover, although many studies have shed light on gut microbiome in health and disease, and established correlations with various diseases in both experimental animals and humans, the causal relationship and molecular mechanisms remain unclear in the most studies. Besides, the application strategies and safety problems in gut microbiome interventions need to be taken into account ( Swann et al., 2020 ). With the biotechnological and computational advancement in this field, more and further explorations will certainly be conducted.
Currently, the volume of scientific literatures about the gut microbiome and disease presents exponential growth. Although there are narrative reviews of gut microbiome and a specific disease, the entire research filed of the gut microbiome and disease is still lack of systematic and quantitative analysis. It is essential to outline this research domain to provide relevant scholars a ready and holistic grasp. Bibliometrics is a multidisciplinary discipline of quantitative analysis of all knowledge carriers by mathematical and statistical methods ( Yu et al., 2018 ). The number and citations of academic publications can reflect the knowledge structure and development features of a scientific domain. Bibliometric analysis is beneficial for identifying and mapping the cumulative scientific knowledge and evolutionary nuances of scientific fields ( Donthu et al., 2021 ). Bibliometrics has been widely used in many other fields, such as economic management, information science, energy and environment ( Yu et al., 2020b ). Therefore, we profile the research landscape of gut microbiome and disease with bibliometric methodology, to provide historical context and detect hot topics and emerging areas in this field. Furthermore, future evolutionary paths and challenges in this field are discussed.
Materials and methods
Data source and search strategy.
Data were retrieved by an advanced search from the WoSCC of Clarivate Analytics, 1 a curated collection of high-quality academic material on the Web of Science™ platform generally used for literature search, journal selection, research evaluation and bibliometric analysis ( Li et al., 2018 ). To avoid bias due to daily updates of the database, document retrieval and export were performed within a single day (May 1, 2022). In order to include as far as possible relevant publications, synonyms for the gut microbiota and disease were included in the search strategy, and the boolean search was set to TS = [(gut* OR intestin* OR gastrointestin* OR gastro-intestin*) AND (microbiota OR microbiome OR flora OR microflora OR bacteria OR microbe* OR microorganism*)] AND TS = (disease*). The time span of publications was set as 1985-01-01 to 2021-12-31. The full record and cited references of the retrieved documents were saved for further analysis. The workflow of the study was presented in Supplementary Figure 1 .
Bibliometric analysis and data visualization
Given that original research is considered as primary literature and presents new knowledge to a certain research area, the “Articles” type of documents was used to evaluate the trends and hotspots of the gut microbiome and disease research. Citespace ( Chen et al., 2012 ; v5.8.R3) was used to analyze reference co-citation, keyword co-occurrence, keywords burst and cooperation relationships among countries, institutions and authors. The Gephi ( Bastian et al., 2009 ; v.0.9.2) was used to construct network graphs.
Research trend of gut microbiome in human health and disease
The increase of publications number and subject categories.
A total of 45,207 academic publications were retrieved from WoSCC, and publication years were distributed from 1996 to 2021. Among these publications, articles account for 66.074% (29,870 records), reviews account for 29.445% (13,311 records), other document types and their percentages see Supplementary Table 1 . The overall output of publications has increased approximately exponentially for the last two decades ( Figure 1A ). Most of the studies were reported in the recent 15 years (n = 27,558, 92.260%). A turning point can be observed around 2007 ( Figures 1B , ,C), C ), since that, the number of publications has been rising drastically. This is partially because of the invention of next-generation sequencing technologies. Other important reasons are the completion of the Human Genome Project (HGP) the launch of the Human Microbiome Project (HMP) and the Metagenomics of The Human Intestinal Tract (MetaHIT). The number of articles supported by fund(s), funding agencies, and funding projects has also been increasing for 26 years ( Figures 1C , ,D), D ), and the percentage of articles supported by fund(s) has been up to 80% in recent 5 years. These results reveal that the gut microbiome and disease research is active and has received increasing attention.

The trend of publications and funding in the research field of gut microbiome and disease. (A) The cumulative number of publications in each year and their exponential regressions. (B) The year-on-year growth rate of publications. (C) The number and percentage of funded articles. (D) The number of funding agencies and projects each year.
A variety of web of science categories (174/254) are involved in these published articles (29,870), and the number was gradually ascending to 130 in 2021 ( Supplementary Figure 2A ), which suggests that the scientific field presents interdisciplinary characteristics ( Supplementary Figures 3 , 4A – D ). The top 10 subject categories are Microbiology, Immunology, Multidisciplinary Sciences, Gastroenterology & Hepatology, Biochemistry & Molecular Biology, Nutrition & Dietetics, Food Science & Technology, Pharmacology & Pharmacy and Biotechnology & Applied Microbiology, and the co-occurrence network of subject categories in the recent 5 years is shown in Supplementary Figure 2B . This research area shows tight relationships with medicine, immunology and nutrition besides microbiology ( Supplementary Figures 2C , 5 ). There is remarkable growth in the number of articles related to cancer and the nerve system every year ( Supplementary Figures 2D , 5 ).
The shift of research topics
The top 200 out of 33,664 keywords by frequency in 1996–2021 were used to construct heatmaps. These keywords were classified into nine categories, including “Definition,” “Technology,” “Experimental subjects” ( Supplementary Figure 6 ), “Diseases/Conditions,” “Immunity,” “Mechanism,” “Metabolism,” “Intervention,” and “Microbes” ( Figure 2 ). Description about this scientific area shifts gradually from “microflora” to “microbiota” and “microbiome” ( Supplementary Figure 6A ). A technological transition from PCR to sequencing and omics technologies is detected ( Supplementary Figure 6B ). The primary research subjects include “child,” “infant,” “pregnancy,” and “mice” ( Supplementary Figure 6C ).

Heatmap of the top 200 keywords by frequency from articles published in 1998–2021. (A) The keywords related to “Diseases/Conditions.” (B) The keywords related to “Immunity,” “Mechanism,” and “Metabolism.” (C) The keywords related to “Intervention.” (D) The keywords related to “Microbes.”
Intestinal diseases have higher keywords frequency than others ( Figure 2A ). Additionally, most intestinal diseases cover almost the whole period in this research area, and part of them remain hot topics with high keywords frequency, such as “Inflammatory bowel disease,” “ulcerative colitis,” and “Crohn’s disease.” This is easy to understand, considering that the intestines provide a natural habitat for these microorganisms and exchange substances with them. While extra-intestinal diseases draw scientists’ attention in the later years, such as obesity, diabetes, Alzheimer’s disease, Parkinson’s disease, cardiovascular disease, hypertension and depression. Due to the COVID-19 pandemic, the connection between it and the gut microbiota was also established ( Figure 2A ). Metabolism-related topics with the highest focus are short chain fatty acids (SCFA), butyrate, bile acid and trimethylamine N-oxide (TMAO). Hot topics related to immunity are cytokines, innate immunity and intestinal barrier ( Figure 2B ). Probiotics, antibiotics, diet and prebiotics are popular topics in invention of gut microbiome, while FMT and high fat diet are emerging topic ( Figure 2C ). In this field, the primary concern of microbes are probiotics and intestinal pathogens ( Figure 2D ). Supplementary Figure 7 shows the changing trend in the top 15 keywords over time.
Keywords burst means the sudden increase of keywords frequency in a specific period, which involves two attributes—burst strength and duration. A total of 726 keywords were detected as burst keywords. These keywords were also classified into seven categories, i.e., “Definition,” “Technology” ( Supplementary Figure 8 ), “Diseases/Conditions,” “Metabolism,” “Immunity,” “Mechanism,” and “Intervention” ( Figure 3 ). Description and technological shift in the development of the field are also observed ( Supplementary Figure 8 ). 16S rRNA sequencing has become the most useful and active technique to decipher the diversity and abundance of the microbiome.

Keywords with strongest bursts from 1998 to 2021. (A) The keywords related to “Diseases/Conditions.” (B) The keywords related to “Immunity,” “Mechanism,” and “Metabolism.” (C) The keywords related to “Intervention.” Asterisk (*) indicate the origin words missed single quotation marks or blank and has been corrected. The red bars indicate burst duration and strength.
The top 5 burst keywords related to disease with the highest burst strength are “Crohn’s disease,” “dysbiosis,” “atopic disease,” “ulcerative colitis,” and “Parkinson’s disease.” “Intestinal inflammation” has the longest burst duration (1998–2018) followed by “Crohn’s disease” and “diarrhea.” Overall burst keywords related to intestinal disease covered the early and middle period (−2013) such as “enterocolitis,” “Crohn’s disease,” and “diarrhea”; while extra-intestinal diseases take up the later period (2014–2021) such as “obesity,” “cardiovascular disease,” “Alzheimer’s disease,” “anxiety,” “Parkinson’s disease,” “dementia,” “depression,” “hypertension,” and “type 2 diabetes mellitus” ( Figure 3A ). The burst keywords involving “Immunity,” “Mechanism,” and “Metabolism” are presented in Figure 3B . Among them “colonic fermentation,” “bile,” and “lipopolysaccharide” burst at early period. On the contrary “SCFA,” “TMAO,” and “phosphatidylcholine” are detected as burst keywords in recent years. As for the intervention of the gut microbiome “FMT,” “fiber,” “dietary supplementation” and high-fat diet are identified as burst keywords over the last several years ( Figure 3C ).
Knowledge map of gut microbiome and disease
Co-cited references are those articles cited together by other articles, and thus, can be regarded as the knowledge basis of a certain field. The knowledge map of the co-occurrence references reveals the developments and characteristics of this field ( Figure 4 ). The nodes size, i.e., co-citations times, is generally larger than the previous one since 2007. The largest component of the co-citation network is divided into 41 clusters (size >1), which show the diversity of research topics. The top 10 articles by cited times and co-cited times are listed in Supplementary Tables 2 , 3 , respectively. A total of 2,115 articles are detected as citation bust, the highest strength is 185.33, and the longest duration is 9 years. Articles with high centrality are often considered as critical points or turning points in a field, and the top 10 articles are marked in Figure 4 and listed in Supplementary Table 4 ; their publication time range from 2002 to 2010.

The top 5 largest components of co-citation network on the gut microbiome and disease between 1997 and 2021. Each node represents a cited article, and the size reflects the number of co-citations, and the edges denote the co-cited relationships among articles.
Present status of scientific collaboration and journal analysis
Country cooperation.
The data of publications in recent 5 years is utilized to evaluate the present cooperative status in the research filed of the gut microbiome and disease. A total of 18,049 articles were from 157 countries/regions in 2017–2021; the top 10 countries in terms of publications and centrality are shown in Supplementary Table 5 . More than half of the publications were produced by the United States ( n = 5,323) and China ( n = 5,253), accounting for 29.5 and 29.1% of the total, respectively, while every other country contributed less than 6% of the total. Figure 5A shows the international research collaborations among the leading countries in papers output in this field. A higher centrality indicates that more information is passed through the node, which implies the importance of nodes in the network. The United States has the highest centrality value (0.52), followed by England (0.31) and Germany (0.14). Besides, the United States is the most active nation with the largest number of publications in this research filed. Although China’s publications amount is commensurate with the United States, it lagged behind in collaborations with other countries. Japan and India also had poor performance in collaborations among these top 15 countries.

The cooperation network in different levels from 2017 to 2021. (A) The cooperation network of the top 15 most productive countries. The colored rings in the node represent publications amount in different years. The lines’ thickness and color indicate the strength of cooperation relationships and the year of first cooperation, respectively. (B) The largest component of cooperation network of institutions. The top 10 institutions in the number of publications are colored. (C) The largest component of author cooperation network. The top 10 authors in the number of publications are colored. The nodes’ size and the thickness of the lines positively correlated to the production of papers and the strength of cooperation relationships, respectively.
Institution cooperation
There are 559 out of 12,186 institutions that participated in the publication of more than 25 articles from 2017 to 2021. The top 10 institutions in terms of publications and centrality are listed in Supplementary Table 6 . Harvard Medical School had the largest number of publications ( n = 329) among institutions worldwide, followed by the Chinese Academy of Sciences ( n = 305) and the University of California San Diego ( n = 225). The institutions cooperation network is shown in Figure 5B . The Harvard Medical School and the University of California San Diego are active institutions in this research filed in both publications and cooperation. There an obvious inner-country cooperation trend in the institutions cooperation network, especially in United States and China due to their large number of publications. But institutions cooperation network in the United States is more intensive than in China.
Author cooperation
Up to 79,972 authors were involved in the publication of the 18,049 articles from2017 to 2021, and a total of 218 authors participated in the publication of at least 25 articles. Detailed information on the top 10 authors in terms of publications and centrality is provided in Supplementary Table 7 . Rob Knight is the most prolific author in the field of gut microbiome and disease, followed by Wei Chen and Hao Zhang. The author cooperation network is shown in Figure 5C . Similar to the institutions cooperation network, the inner-country cooperation pattern is also observed in the author cooperation network.
Journal analysis
114 out of 2,720 journals published more than 25 articles on the gut microbiome and disease over the 5 years. As shown in Supplementary Table 8 , the top 20 journals with the highest number of articles included 4,356 records, which accounts for 24.13% of the total. Scientific reports are the most productive journal, with 605 articles in this field, followed by Frontiers in microbiology (510) and Plos one (409). Although Gut ranks 19th in terms of the number of articles published, it has the highest IF (23.059) among the 20 journals, followed by Nature communications (22.059) and Microbiome (14.650), and Gut is the most-cited journal with 92.879 citations per article. There was a significant positive correlation between impact factor values and the citations per article (R 2 = 0.869, p < 0.001) for the top 20 most productive journals ( Supplementary Figure 9 ).
Hot topics and emerging trend
By combining Figure 2 with Figure 3 , we can see that diseases that have attracted continuous attention are IBD, UC and CD. In contrast, other diseases have come into researchers’ notice in recent years, such as obesity, dysbiosis, diabetes, cardiovascular disease, Alzheimer’s disease, Parkinson’s disease, hypertension, depression and COVID-19. Compared to diet/nutrition and drugs, probiotics draw more attention, while FMT can be identified as a frontier of research.
The data of publications in the recent 5 years is used to assess the current research status of gut microbiome and disease. The timeline view of keywords co-occurrence network reveals the development of gut microbiome and disease ( Supplementary Figure 10 ). This network is divided into 21 clusters, which present the major subtopics in this field. Except “#0 growth performance” and “#14 oral microbiome” is irrelevant, other can be consider ongoing topics in recent. Campylobacter jejuni is commonly found in animal feces and causes human gastroenteritis, but the average year of “#16 campylobacter jejuni” is older than other clusters. In addition to utilizing keywords, co-cited references are also used to detect research hotspots and emerging trends. A total of 13 clusters are identified in the co-cited references network, and each cluster corresponds to a line of research ( Figure 6 ). Except “#7 Aquaculture”, other clusters closely related to this field. Among these clusters, “#0 Inflammatory bowel disease” contains most of the nodes, which means that it has been widely reported. “#1 Metatrascriptomics” has most of the citation burst articles, followed by “#5 Multiple sclerosis,” “#3 trimethylamine N-oxide,” “#2 Parkinson’s disease,” which indicate they are active research areas.

Co-cited references network from 2017 to 2021. Each node represents a cited article.
Diseases related to gut microbiome
A total of 541 diseases and conditions are retrieved from the Centers for Disease Control and Prevention (CDC, https://www.cdc.gov/DiseasesConditions/ ) and the Illinois Department of Public Health (IDPH, https://dph.illinois.gov/topics-services/diseases-and-conditions.html ), and they are used to search against WoSCC to depict research status of the gut microbiome with them. 73 diseases and conditions have more than 100 records in WoSCC from 1996 to 2021, and their publications trend are visualized in Figure 7 . “Overweight and Obesity” is an area of focus, possessing the largest number of articles. While, “Stress” ranked second, possibly because of its lexical ambiguity and irrelevant articles are hit. Gut-related diseases have been more reported than others, which are consistent with previous results. There are 5,984 records related to gut microbiome and cancers, and 19 types of cancers are involved. Colorectal (Colon) Cancer is in the first echelon with the largest amount of records ( n = 2,489), and the second echelon includes Breast Cancer, Prostate Cancer, Lung Cancer, Pancreatic Cancer, Leukemia and Liver Cancer, and others belongs to the third echelon with records less than 50 ( Supplementary Figure 11 ).

Publication trend of diseases and conditions related to gut microbiome from 1998 to 2021.
Due to limitation of database, the earliest document is in 1996 in this study. But the first publication in this filed actually could trace back to 1958, when Eiseman et al. reported the successful treatment of pseudomembranous enterocolitis using a faecal enema ( Eiseman et al., 1958 ). Studies about the gut microbiome and disease have increased tremendously over the last decades and present exponential growth, which revealed the important role of the gut microbiome in human health and disease ( Gebrayel et al., 2022 ). Given that there are many unknowns about the gut microbiome and their potential applications in the prevention, diagnosis and therapy of diseases, this research scope will continue to attract keen interest among scientists, and further explorations will be conducted in the future.
To date, numerous studies have indicated that the intestinal microbiome is associated with various diseases, particularly digestive tract diseases ( Nouvenne et al., 2018 ). However, most of them were observational (i.e., different in diversity, taxa, OTU and functions among groups) and did not reveal cause and effect ( Koh and Bäckhed, 2020 ; Walter et al., 2020 ). It’s necessary to rethink whether there are causal relationships and whether the microbiota is a dominant or a crucial driving factor when surveying gut microbiome in diseases. Metabolites play an essential role in interactions between microbes and host cells, the altered composition of microbes could bring about a cascading impact on the immune system, and then effect the host health status ( Rooks and Garrett, 2016 ). Currently, the most extensively studied metabolites are SCFA, bile acids, TMAO, and amino acid-derived metabolites ( Liu et al., 2022 ), and other microbial metabolites such as lipids ( Schoeler and Caesar, 2019 ), carbohydrates ( Cheng et al., 2020 ) have also been proved to be essential for microbe-host interaction. However, comprehensive mechanisms that explain the link between the gut microbiome and most diseases remain poorly understood. Therefore, we encourage researchers to generate hypotheses based on observed differences in taxa and functions, and to independently validate it whenever possible.
The recent development of multi-omics approaches, such as metataxonomics [16S rRNA and ITS (Internal Transcribed Spacer) sequencing], shotgun metagenomics, metatranscriptomics, metaproteomics, and metabolomics, has enabled efficient characterization of microbial communities. These techniques not only provide the taxonomic profile of the microbial community but also assess their latent functions and metabolic activities ( Zhang et al., 2019 ). The biomarkers detected by these -omics technologies could help to elucidate potential mechanisms of these commensals in health and disease ( Lloyd-Price et al., 2019 ; Zhou et al., 2019 ; Mars et al., 2020 ). However, this also brings challenges to multi-omics data integration and mining ( Whon et al., 2021 ). Currently, methods of data integration include two categories, i.e., multi-staged analysis and meta-analysis. Multi-staged integration means using two or more categorical features of the data. For example, metagenomics is combined with metabolomics ( Oh et al., 2020 ). Meta-analysis attempts to systematically merge data across multiple studies and transform it into metadata that can be analyzed simultaneously ( Armour et al., 2019 ; Wang et al., 2021 ; Drewes et al., 2022 ), which reduce study bias, increase statistical power and improve overall biological understanding of a study effect. As for data mining, machine learning has been applied to find biomarkers and carry out classification or prediction tasks, such as diagnosis, disease course, and disease severity ( Marcos-Zambrano et al., 2021 ). But its limitation is requiring large amounts of data and lacking of interpretability. There are platforms and tools developed for multi-omics data integrating and mining, such as Qiita ( Gonzalez et al., 2018 ), MicrobiomeAnalyst ( Dhariwal et al., 2017 ), NetMoss ( Xiao et al., 2022 ), tmap ( Liao et al., 2019 ), which may aid in understanding the correlation between the gut microbiome and disease.
Besides investigating the relationship and mechanisms between the gut microbiome and diseases, it also is an interesting subject to modulate the gut microbiota to benefit health and reduce the risk of diseases. The main intervention strategies include diet/nutrition, dietary supplement, medicine and FMT. Diet is a feasible and easy measure to maintain homeostasis or increase the diversity of the gut microbiota. The question is, what type of diet can help to establish a good and stable intestinal microbiota ( Leeming et al., 2019 ). Previous studies have indicated that FMT could restore gut microbial diversity and eliminate Clostridioides difficile infection (CDI; Kelly et al., 2021 ), which has encouraged research into the use of FMT for other diseases, such as ulcerative colitis and Crohn’s disease. While, results of FMT are not always desirable and the effectiveness is highly variable ( Nie et al., 2019 ). It is assumed that the beneficial functions of therapeutic microbes are based on colonization and retention in sufficient quantity for enough time in recipients ( Lee et al., 2017 ; Chu et al., 2021 ). Therefore, the selection of appropriate donors or its microbes and efficient colonization plays an essential role in patient response ( Woodworth et al., 2017 ; Jouhten et al., 2020 ). It is also important to take into consideration how to appropriately evaluate the safety and efficacy for a given intervention ( Green et al., 2020 ; Haifer et al., 2021 ). It is possible and valuable to develop novel diagnostic, prognostic and therapeutic strategies based on microbiome manipulation. The management of common diseases could be transformed by translating microbiome research into treatments that regulate the microbiome. Although there are some microbiome interventions as effective treatment for improving health conditions, its detail mechanisms are not fully understood.
As one of the hot topics in gut microbiome and diseases, IBD is a chronic inflammatory gut pathological condition, and represented by CD and UC. Both diseases are characterized by diarrhea, rectal bleeding, abdominal pain, fatigue and weight loss, but differentiate in clinical manifestations of inflammation and intestinal localization ( Le Berre et al., 2020 ). Although a complete understanding of IBD pathogenesis is unclear, various risk factors associated with IBD have been identified, such as host genetic susceptibility, environmental variables, immune response and gut microbiome ( Chang, 2020 ). Indeed, studies in human subjects have shown that the gut microbiome is significant different in patients with IBD compared with that in healthy individuals ( Halfvarson et al., 2017 ; Lloyd-Price et al., 2019 ), such as reduced species richness and diversity, and lower temporal stability. Among them, the certain microbial taxa that are enriched or depleted in IBD, including bacteria, archaea, fungi, and viruses ( Iliev and Cadwell, 2021 ), is usually interpreted as the imbalance between beneficial and pathogenic microbe, however, the results differ between studies ( Schirmer et al., 2019 ). Alteration of gut microbial metabolites in IBD patients also detected, including fatty acids, amino acids and derivatives and bile acids, which may act as key regulators in the pathogenesis of IBD ( Li et al., 2022 ; Paik et al., 2022 ). Although UC and CD are similar in epidemiologic, immunologic, therapeutic and clinical features, they fell into two distinct groups at the gut microbiome pattern ( Pascal et al., 2017 ). The shifts in gut microbial community have been proven to be potential as diagnostic biomarkers of IBD ( Zhou et al., 2018 ; Guo et al., 2022 ), which could be used to develop non-invasive diagnostic or monitor methods, while independent external validation is necessary before it can be used in clinic. There are therapeutic advances in gut microbiome modulation in patients with IBD, and a variety of microbiome-modulating interventions are proposed for treatment, such as probiotics, prebiotics, antibiotics, FMT, and dietary supplements ( Eindor-Abarbanel et al., 2021 ). However, retrospective studies and meta-analyses on antibiotic use in UC and CD and long-term outcomes are controversial ( Ledder, 2019 ). Similarly, the use of probiotics for the effective treatment of IBD remains inconclusive ( Zhao et al., 2018 ). Due to the complexity and variety of IBD pathogenesis, personalized and multidimensional treatment will likely be required where microbiome-modulating therapy is coupled with other therapies. Changes in the gut microbiome seemed to play an important role in the onset of IBD, yet longitudinal studies of the gut microbiome are needed to move from association toward causation and modulation.
The research of the gut microbiome in human health and disease remains loaded with challenges. Gut microbiota is a complex and dynamic consortium influenced by multiple factors ( Spencer et al., 2019 ; Kurilshikov et al., 2021 ; Gacesa et al., 2022 ). Changes in hosts’ lifestyle, such as diet, medication use, age, and socioeconomic status can lead to data reproducibility problems and statistical underpower. Recruiting participants with well-defined disease or at-risk conditions and well data management is important to reduce background noise. In addition, relatively few controlled samples in the trial may cause inconsistent results in the same disease. Because of the need for long longitudinal study, the influence from sample collection and storage and batch effects need to be avoided ( Wang and LêCao, 2020 ; Poulsen Casper et al., 2021 ). Nowadays, gut microbiome research involves multi-disciplinary, not only microbiology and gastroenterology but also bioinformatics, mathematics, biochemistry, immunology and ecology, which pose challenges for single researcher ( Mirzayi et al., 2021 ). There are gaps in scientific and technological power among countries, United States has established its leadership in this field. Therefore, we propose to enhance coordination and collaboration across the field among scientific communities to tackle shared challenges and explore new frontiers jointly. At present, inner-country cooperation pattern was observed at the institution and author levels, while a dynamic analysis of the collaboration networks based on different periods can show the evolution of collaborated patterns ( Yu et al., 2020a ). Effective international cooperation could promote academic exchanges. It may be a solution to the research of gut microbiome in disease by conducting well-designed large-scale cohort studies and randomized clinical trials, meanwhile combining multi-omics techniques and integrating microbiome data ( Heintz-Buschart et al., 2016 ; Park et al., 2022 ). Due to confounding factors, it is necessary to establish standardized experimental procedures and subsequent data analysis pipelines ( Szóstak et al., 2022 ). While experimental animal models can provide fascinating insights into the role of the microbiome in disease states, they rarely recapitulate the complete human phenotype ( Hugenholtz and de Vos, 2018 ; Kieser et al., 2022 ). Therefore, extrapolations to human diseases have to be viewed with caution, and more rigorous experiments are required. The current focus concerning gut microbiota is mainly on bacteria, which neglects the significance of microbial intra- and inter-kingdom interaction. Fungi and viruses also impact the gut microbiota and host ( van Tilburg Bernardes et al., 2020 ), although knowledge about their relationship with dysbiosis is limited ( Carding et al., 2017 ; Beller and Matthijnssens, 2019 ). A recent study identified signature fungi in colorectal cancer and adenoma patients from multiple cohorts, and observed trans-kingdom interactions between enteric fungi and bacteria in colorectal cancer progression ( Lin et al., 2022 ).
Bibliometric analysis is increasingly being used to assess hot topics and emerging areas of a specific field. Compared to narrative reviews that provide qualitative summary and commentary of published literature in a field, it quantitatively investigates the status of interdisciplinary fields based on citations and other statistical information regarding publications. In the future, the combination of the two will present a more precise historical context and future trajectory for a field. There are situations that need to be balanced in bibliometric analysis. The first situation is choosing databases. Other databases such as PubMed and Scopus also can be set as the data source, Scopus covers even more journals and also contains citation records. However, Web of Science (WoS) assigns document type labels more accurately than Scopus ( Yeung, 2019 ), and we only filtered for original articles for the downstream analysis. The second situation is setting a search strategy. Well-defined search terms should include publications related to the field and exclude irrelevant ones as far as possible. It seems to be inevitable to contain irrelevant publications except for manual verification, but we believe that it is reliable to reflect the global trend and hot topics by these multi-aspect analysis. Artificial intelligence technology has the potential to realize semantic detection of publications and determine whether they belong to a specific theme. This would be especially useful for bibliometric analysis with massive volume of data and improve the accuracy of results.
Bibliometric methods are quantitative by nature to examine unlimited quantities of publications. But our study also comes with certain limitations. Firstly, due to the nature of the bibliometric methodology, the relationship between some bibliometric metrics and their assertions about research quality is often unclear ( Wallin, 2005 ). Secondly, our study only retrieved data from WoS, yet a combination with other databases can be performed in similar type of research. Thirdly, synonymous words need be merged together during the analysis.
In conclusion, based on the detailed bibliometrics analysis of gut microbiome and disease, we present a comprehensive overview of this evolving subject over the past 26 years. These results indicate that gut microbiome and disease is an active research field, and publications on this subject have proliferated over the past decades. The current research mainly focuses on gastrointestinal diseases, while extra-intestinal diseases are also rising, such as nerve-related diseases. Although extensive correlative studies have been performed, the molecular mechanisms still need to be explored. Overall, gut microbiome research shows a multitude of challenges and great opportunities.
Data availability statement
Author contributions.
All authors contributed to the study conception and design. Material preparation, data collection and analysis were performed by ZH, KL, and WM. The first draft of the manuscript was written by ZH and all authors commented on previous versions of the manuscript. All authors contributed to the article and approved the submitted version.
This project was supported by Science and Technology innovation Plan of Shanghai (19391902000).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to acknowledge the tremendous work and effort of our research team and we are deeply grateful for their constant support.
1 https://clarivate.com/
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.1018594/full#supplementary-material
- Almeida A., Mitchell A. L., Boland M., Forster S. C., Gloor G. B., Tarkowska A., et al.. (2019). A new genomic blueprint of the human gut microbiota . Nature 568 , 499–504. doi: 10.1038/s41586-019-0965-1, PMID: [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Armour C. R., Nayfach S., Pollard K. S., Sharpton T. J. (2019). A metagenomic meta-analysis reveals functional signatures of health and disease in the human gut microbiome . mSystems 4 , e00332–e00318. doi: 10.1128/mSystems.00332-18, PMID: [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Bastian M., Heymann S., Jacomy M. (2009). Gephi: an open source software for exploring and manipulating networks . ICWSM 3 , 361–362. doi: 10.1609/icwsm.v3i1.13937 [ CrossRef ] [ Google Scholar ]
- Beller L., Matthijnssens J. (2019). What is (not) known about the dynamics of the human gut virome in health and disease . Curr. Opin. Virol. 37 , 52–57. doi: 10.1016/j.coviro.2019.05.013, PMID: [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Carding S. R., Davis N., Hoyles L. (2017). Tthe human intestinal virome in health and disease . Aliment. Pharm. Ther. 46 , 800–815. doi: 10.1111/apt.14280, PMID: [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Chang J. T. (2020). Pathophysiology of inflammatory bowel diseases . New Engl. J. Med. 383 , 2652–2664. doi: 10.1056/NEJMra2002697 [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Chen C., Hu Z., Liu S., Tseng H. (2012). Emerging trends in regenerative medicine: a scientometric analysis in CiteSpace . Expert Opin. Biol. Ther. 12 , 593–608. doi: 10.1517/14712598.2012.674507, PMID: [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Cheng X., Zheng J., Lin A., Xia H., Zhang Z., Gao Q., et al.. (2020). A review: roles of carbohydrates in human diseases through regulation of imbalanced intestinal microbiota . J. Funct. Foods 74 :104197. doi: 10.1016/j.jff.2020.104197 [ CrossRef ] [ Google Scholar ]
- Chu N. D., Crothers J. W., Nguyen L. T., Kearney S. M., Smith M. B., Kassam Z., et al.. (2021). Dynamic colonization of microbes and their functions after fecal microbiota transplantation for inflammatory bowel disease . mBio 12 , e00975–e00921. doi: 10.1128/mBio.00975-21 [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Coyte K. Z., Rakoff-Nahoum S. (2019). Understanding competition and cooperation within the mammalian gut microbiome . Curr. Biol. 29 , R538–R544. doi: 10.1016/j.cub.2019.04.017, PMID: [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Dhariwal A., Chong J., Habib S., King I. L., Agellon L. B., Xia J. (2017). MicrobiomeAnalyst: a web-based tool for comprehensive statistical, visual and meta-analysis of microbiome data . Nucleic Acids Res. 45 , W180–W188. doi: 10.1093/nar/gkx295, PMID: [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Dinan T. G., Cryan J. F. (2017). The microbiome-gut-brain axis in health and disease . Gastroenterol. Clin. 46 , 77–89. doi: 10.1016/j.gtc.2016.09.007 [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Donthu N., Kumar S., Mukherjee D., Pandey N., Lim W. M. (2021). How to conduct a bibliometric analysis: an overview and guidelines . J. Bus. Res. 133 , 285–296. doi: 10.1016/j.jbusres.2021.04.070 [ CrossRef ] [ Google Scholar ]
- Drewes J. L., Chen J., Markham N. O., Knippel R. J., Domingue J. C., Tam A. J., et al.. (2022). Human colon cancer-derived Clostridioides difficile strains drive colonic tumorigenesis in mice . Cancer Discov. 12 :1873. doi: 10.1158/2159-8290.CD-21-1273, PMID: [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Durack J., Lynch S. V. (2018). The gut microbiome: relationships with disease and opportunities for therapy . J. Exp. Med. 216 , 20–40. doi: 10.1084/jem.20180448, PMID: [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Eindor-Abarbanel A., Healey G. R., Jacobson K. (2021). Therapeutic advances in gut microbiome modulation in patients with inflammatory bowel disease from pediatrics to adulthood . Int. J. Mol. Sci. 22 :12506. doi: 10.3390/ijms222212506, PMID: [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Eiseman B., Silen W., Bascom G. S., Kauvar A. J. (1958). Fecal enema as an adjunct in the treatment of pseudomembranous enterocolitis . Surgery 44 , 854–859. [ PubMed ] [ Google Scholar ]
- Fei N., Zhao L. (2013). An opportunistic pathogen isolated from the gut of an obese human causes obesity in germfree mice . ISME J. 7 , 880–884. doi: 10.1038/ismej.2012.153, PMID: [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Gacesa R., Kurilshikov A., Vich Vila A., Sinha T., Klaassen M. A. Y., Bolte L. A., et al.. (2022). Environmental factors shaping the gut microbiome in a Dutch population . Nature 604 , 732–739. doi: 10.1038/s41586-022-04567-7, PMID: [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Gebrayel P., Nicco C., Al Khodor S., Bilinski J., Caselli E., Comelli E. M., et al.. (2022). Microbiota medicine: towards clinical revolution . J. Transl. Med. 20 :111. doi: 10.1186/s12967-022-03296-9, PMID: [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Gonzalez A., Navas-Molina J. A., Kosciolek T., Mcdonald D., Vázquez-Baeza Y., Ackermann G., et al.. (2018). Qiita: rapid, web-enabled microbiome meta-analysis . Nat. Methods 15 , 796–798. doi: 10.1038/s41592-018-0141-9, PMID: [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Gopalakrishnan V., Helmink B. A., Spencer C. N., Reuben A., Wargo J. A. (2018). The influence of the gut microbiome on cancer, immunity, and cancer immunotherapy . Cancer Cell 33 , 570–580. doi: 10.1016/j.ccell.2018.03.015, PMID: [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Green J. E., Davis J. A., Berk M., Hair C., Loughman A., Castle D., et al.. (2020). Efficacy and safety of fecal microbiota transplantation for the treatment of diseases other than Clostridium difficile infection: a systematic review and meta-analysis . Gut Microbes 12 , 1854640–1854625. doi: 10.1080/19490976.2020.1854640, PMID: [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Grice E. A., Segre J. A. (2012). The human microbiome: our second genome . Annu. Rev. Genomics Hum. Genet. 13 , 151–170. doi: 10.1146/annurev-genom-090711-163814, PMID: [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Guo X., Huang C., Xu J., Xu H., Liu L., Zhao H., et al.. (2022). Gut microbiota is a potential biomarker in inflammatory bowel disease . Front. Nutr. 8 :818902. doi: 10.3389/fnut.2021.818902, PMID: [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Haifer C., Saikal A., Paramsothy R., Kaakoush N. O., Leong R. W., Borody T. J., et al.. (2021). Response to faecal microbiota transplantation in ulcerative colitis is not sustained long term following induction therapy . Gut 70 , 2210–2211. doi: 10.1136/gutjnl-2020-323581 [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Halfvarson J., Brislawn C. J., Lamendella R., Vázquez-Baeza Y., Walters W. A., Bramer L. M., et al.. (2017). Dynamics of the human gut microbiome in inflammatory bowel disease . Nat. Microbiol. 2 :17004. doi: 10.1038/nmicrobiol.2017.4, PMID: [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Heintz-Buschart A., May P., Laczny C. C., Lebrun L. A., Bellora C., Krishna A., et al.. (2016). Integrated multi-omics of the human gut microbiome in a case study of familial type 1 diabetes . Nat. Microbiol. 2 :16180. doi: 10.1038/nmicrobiol.2016.180 [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Heintz-Buschart A., Wilmes P. (2018). Human gut microbiome: function matters . Trends Microbiol. 26 , 563–574. doi: 10.1016/j.tim.2017.11.002, PMID: [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Hugenholtz F., De Vos W. M. (2018). Mouse models for human intestinal microbiota research: a critical evaluation . Cell. Mol. Life Sci. 75 , 149–160. doi: 10.1007/s00018-017-2693-8, PMID: [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Iliev I. D., Cadwell K. (2021). Effects of intestinal fungi and viruses on immune responses and inflammatory bowel diseases . Gastroenterology 160 , 1050–1066. doi: 10.1053/j.gastro.2020.06.100, PMID: [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Jiao J. Y., Liu L., Hua Z. S., Fang B. Z., Zhou E. M., Salam N., et al.. (2021). Microbial dark matter coming to light: challenges and opportunities . Natl. Sci. Rev. 8 :nwaa280. doi: 10.1093/nsr/nwaa280, PMID: [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Jouhten H., Ronkainen A., Aakko J., Salminen S., Mattila E., Arkkila P., et al.. (2020). Cultivation and genomics prove long-term colonization of Donor’s Bifidobacteria in recurrent Clostridioides difficile patients treated with fecal microbiota transplantation . Front. Microbiol. 11 :1663. doi: 10.3389/fmicb.2020.01663, PMID: [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Kelly C. R., Yen E. F., Grinspan A. M., Kahn S. A., Atreja A., Lewis J. D., et al.. (2021). Fecal microbiota transplantation is highly effective in real-world practice: initial results from the FMT National Registry . Gastroenterology 160 , 183–192.e3. doi: 10.1053/j.gastro.2020.09.038, PMID: [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Kieser S., Zdobnov E. M., Trajkovski M. (2022). Comprehensive mouse microbiota genome catalog reveals major difference to its human counterpart . PLoS Comput. Biol. 18 :e1009947. doi: 10.1371/journal.pcbi.1009947, PMID: [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Koenig J. E., Spor A., Scalfone N., Fricker A. D., Stombaugh J., Knight R., et al.. (2011). Succession of microbial consortia in the developing infant gut microbiome . Proc. Natl. Acad. Sci. U.S.A. 108 , 4578–4585. doi: 10.1073/pnas.1000081107, PMID: [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Koh A., Bäckhed F. (2020). From association to causality: the role of the gut microbiota and its functional products on host metabolism . Mol. Cell 78 , 584–596. doi: 10.1016/j.molcel.2020.03.005, PMID: [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Krautkramer K. A., Fan J., Bäckhed F. (2021). Gut microbial metabolites as multi-kingdom intermediates . Nat. Rev. Microbiol. 19 , 77–94. doi: 10.1038/s41579-020-0438-4, PMID: [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Kundu P., Blacher E., Elinav E., Pettersson S. (2017). Our gut microbiome: the evolving inner self . Cells 171 , 1481–1493. doi: 10.1016/j.cell.2017.11.024, PMID: [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Kurilshikov A., Medina-Gomez C., Bacigalupe R., Radjabzadeh D., Wang J., Demirkan A., et al.. (2021). Large-scale association analyses identify host factors influencing human gut microbiome composition . Nat. Genet. 53 , 156–165. doi: 10.1038/s41588-020-00763-1, PMID: [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Lau W. L., Tran T., Rhee C. M., Kalantar-Zadeh K., Vaziri N. D. (2021). Diabetes and the gut microbiome . Semin. Nephrol. 41 , 104–113. doi: 10.1016/j.semnephrol.2021.03.005 [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Le Berre C., Ananthakrishnan A. N., Danese S., Singh S., Peyrin-Biroulet L. (2020). Ulcerative colitis and Crohn’s disease have similar burden and goals for treatment . Clin. Gastroenterol. Hepatol. 18 , 14–23. doi: 10.1016/j.cgh.2019.07.005, PMID: [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Ledder O. (2019). Antibiotics in inflammatory bowel diseases: do we know what we’re doing? Transl. Pediatr. 8 , 42–55. doi: 10.21037/tp.2018.11.02, PMID: [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Lee S. T. M., Kahn S. A., Delmont T. O., Shaiber A., Esen Ö. C., Hubert N. A., et al.. (2017). Tracking microbial colonization in fecal microbiota transplantation experiments via genome-resolved metagenomics . Microbiome 5 :50. doi: 10.1186/s40168-017-0270-x, PMID: [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Leeming E. R., Johnson A. J., Spector T. D., Le Roy C. I. (2019). Effect of diet on the gut microbiota: rethinking intervention duration . Nutrients 11 :2862. doi: 10.3390/nu11122862, PMID: [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Li K., Rollins J., Yan E. (2018). Web of science use in published research and review papers 1997–2017: a selective, dynamic, cross-domain, content-based analysis . Scientometrics 115 , 1–20. doi: 10.1007/s11192-017-2622-5, PMID: [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Li M., Yang L., Mu C., Sun Y., Gu Y., Chen D., et al.. (2022). Gut microbial metabolome in inflammatory bowel disease: from association to therapeutic perspectives . Comput. Struct. Biotec. 20 , 2402–2414. doi: 10.1016/j.csbj.2022.03.038, PMID: [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Liao T., Wei Y., Luo M., Zhao G. P., Zhou H. (2019). Tmap: an integrative framework based on topological data analysis for population-scale microbiome stratification and association studies . Genome Biol. 20 :293. doi: 10.1186/s13059-019-1871-4, PMID: [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Lin Y., Lau H. C.-H., Liu Y., Kang X., Wang Y., Ting N. L.-N., et al.. (2022). Altered mycobiota signatures and enriched pathogenic Aspergillus rambellii are associated with colorectal cancer based on multi-cohort fecal metagenomic analyses . Gastroenterology 163 , 908–921. doi: 10.1053/j.gastro.2022.06.038, PMID: [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Liu J., Tan Y., Cheng H., Zhang D., Feng W., Peng C. (2022). Functions of gut microbiota metabolites, current status and future perspectives . Aging Dis. 13 , 1106–1126. doi: 10.14336/AD.2022.0104, PMID: [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Lloyd-Price J., Arze C., Ananthakrishnan A. N., Schirmer M., Avila-Pacheco J., Poon T. W., et al.. (2019). Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases . Nature 569 , 655–662. doi: 10.1038/s41586-019-1237-9, PMID: [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Marcos-Zambrano L. J., Karaduzovic-Hadziabdic K., Loncar Turukalo T., Przymus P., Trajkovik V., Aasmets O., et al.. (2021). Applications of machine learning in human microbiome studies: a review on feature selection, biomarker identification, disease prediction and treatment . Front. Microbiol. 12 :634511. doi: 10.3389/fmicb.2021.634511, PMID: [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Mars R., Yang Y., Ward T., Mo H., Priya S., Lekatz H. R., et al.. (2020). Longitudinal multi-omics reveals subset-specific mechanisms underlying irritable bowel syndrome . Cells 182 , 1460–1473.e17. doi: 10.1016/j.cell.2020.08.007, PMID: [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Mirzayi C., Renson A., Furlanello C., Sansone S.-A., Zohra F., Elsafoury S., et al.. (2021). Reporting guidelines for human microbiome research: the STORMS checklist . Nat. Med. 27 , 1885–1892. doi: 10.1038/s41591-021-01552-x, PMID: [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Morgan X. C., Tickle T. L., Sokol H., Gevers D., Devaney K. L., Ward D. V., et al.. (2012). Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment . Genome Biol. 13 :R79. doi: 10.1186/gb-2012-13-9-r79, PMID: [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Nie P., Li Z., Wang Y., Zhang Y., Zhao M., Luo J., et al.. (2019). Gut microbiome interventions in human health and diseases . Med. Res. Rev. 39 , 2286–2313. doi: 10.1002/med.21584 [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Nouvenne A., Ticinesi A., Tana C., Prati B., Catania P., Miraglia C., et al.. (2018). Digestive disorders and intestinal microbiota . Acta Biomater. 89 , 47–51. doi: 10.23750/abm.v89i9-S.7912, PMID: [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Oh T. G., Kim S. M., Caussy C., Fu T., Loomba R. (2020). A universal gut-microbiome-derived signature predicts cirrhosis . Cell Metab. 32 , 901–888. doi: 10.1016/j.cmet.2020.10.015, PMID: [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Paik D., Yao L., Zhang Y., Bae S., D’agostino G. D., Zhang M., et al.. (2022). Human gut bacteria produce ΤΗ17-modulating bile acid metabolites . Nature 603 , 907–912. doi: 10.1038/s41586-022-04480-z, PMID: [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Park C. H., Hong C., Lee A. R., Sung J., Hwang T. H. (2022). Multi-omics reveals microbiome, host gene expression, and immune landscape in gastric carcinogenesis . iScience 25 :103956. doi: 10.1016/j.isci.2022.103956, PMID: [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Pascal V., Pozuelo M., Borruel N., Casellas F., Campos D., Santiago A., et al.. (2017). A microbial signature for Crohn's disease . Gut 66 , 813–822. doi: 10.1136/gutjnl-2016-313235, PMID: [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Pasolli E., Asnicar F., Manara S., Zolfo M., Karcher N., Armanini F., et al.. (2019). Extensive unexplored human microbiome diversity revealed by over 150, 000 genomes from metagenomes spanning age, geography, and lifestyle . Cells 176 , 649–662.e20. doi: 10.1016/j.cell.2019.01.001, PMID: [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Poulsen Casper S., Kaas Rolf S., Aarestrup Frank M., Pamp Sünje J., Claesen J. (2021). Standard sample storage conditions have an impact on inferred microbiome composition and antimicrobial resistance patterns . Microbiol. Spectr. 9 , e0138721–e0101321. doi: 10.1128/Spectrum.01387-21, PMID: [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Rooks M. G., Garrett W. S. (2016). Gut microbiota, metabolites and host immunity . Nat. Rev. Immunol. 16 , 341–352. doi: 10.1038/nri.2016.42, PMID: [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Schirmer M., Garner A., Vlamakis H., Xavier R. J. (2019). Microbial genes and pathways in inflammatory bowel disease . Nat. Rev. Microbiol. 17 , 497–511. doi: 10.1038/s41579-019-0213-6, PMID: [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Schoeler M., Caesar R. (2019). Dietary lipids, gut microbiota and lipid metabolism . Rev. Endocr. Metab. Disord. 20 , 461–472. doi: 10.1007/s11154-019-09512-0, PMID: [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Sender R., Fuchs S., Milo R. (2016). Revised estimates for the number of human and bacteria cells in the body . PLoS Biol. 14 :e1002533. doi: 10.1371/journal.pbio.1002533, PMID: [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Spencer C. N., Gopalakrishnan V., Mcquade J., Andrews M. C., Helmink B., Khan M. A. W., et al.. (2019). The gut microbiome (GM) and immunotherapy response are influenced by host lifestyle factors . Cancer Res. 79 , 2838–2838. doi: 10.1158/1538-7445.AM2019-2838 [ CrossRef ] [ Google Scholar ]
- Stappenbeck T. S., Hooper L. V., Gordon J. I. (2002). Developmental regulation of intestinal angiogenesis by indigenous microbes via Paneth cells . Proc. Natl. Acad. Sci. U. S. A. 99 , 15451–15455. doi: 10.1073/pnas.202604299, PMID: [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Swann J. R., Rajilic-Stojanovic M., Salonen A., Sakwinska O., Gill C., Meynier A., et al.. (2020). Considerations for the design and conduct of human gut microbiota intervention studies relating to foods . Eur. J. Nutr. 59 , 3347–3368. doi: 10.1007/s00394-020-02232-1, PMID: [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Szóstak N., Szymanek A., Havránek J., Tomela K., Rakoczy M., Samelak-Czajka A., et al.. (2022). The standardisation of the approach to metagenomic human gut analysis: from sample collection to microbiome profiling . Sci. Rep. 12 :8470. doi: 10.1038/s41598-022-12037-3, PMID: [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Van Tilburg Bernardes E., Pettersen V. K., Gutierrez M. W., Laforest-Lapointe I., Jendzjowsky N. G., Cavin J.-B., et al.. (2020). Intestinal fungi are causally implicated in microbiome assembly and immune development in mice . Nat. Commun. 11 :2577. doi: 10.1038/s41467-020-16431-1, PMID: [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Wallen Z. D., Stone W. J., Factor S. A., Molho E., Zabetian C. P., Standaert D. G., et al.. (2021). Exploring human-genome gut-microbiome interaction in Parkinson’s disease . NPJ Parkinsons. Dis. 7 :74. doi: 10.1038/s41531-021-00218-2, PMID: [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Wallin J. A. (2005). Bibliometric methods: pitfalls and possibilities . Basic Clin. Pharmacol. 97 , 261–275. doi: 10.1111/j.1742-7843.2005.pto_139.x, PMID: [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Walter J., Armet A. M., Finlay B. B., Shanahan F. (2020). Establishing or exaggerating causality for the gut microbiome: lessons from human microbiota-associated rodents . Cells 180 , 221–232. doi: 10.1016/j.cell.2019.12.025 [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Wang Y., Lêcao K.-A. (2020). Managing batch effects in microbiome data . Brief. Bioinform. 21 , 1954–1970. doi: 10.1093/bib/bbz105, PMID: [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Wang S., Zeng S., Egan M., Cherry P., Strain C., Morais E., et al.. (2021). Metagenomic analysis of mother-infant gut microbiome reveals global distinct and shared microbial signatures . Gut Microbes 13 , 1–24. doi: 10.1080/19490976.2021.1911571, PMID: [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Whon T. W., Shin N.-R., Kim J. Y., Roh S. W. (2021). Omics in gut microbiome analysis . J. Microbiol. 59 , 292–297. doi: 10.1007/s12275-021-1004-0 [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Woodworth M. H., Carpentieri C., Sitchenko K. L., Kraft C. S. (2017). Challenges in fecal donor selection and screening for fecal microbiota transplantation: a review . Gut Microbes 8 , 225–237. doi: 10.1080/19490976.2017.1286006, PMID: [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Wu Y., Jiao N., Zhu R., Zhang Y., Wu D., Wang A.-J., et al.. (2021). Identification of microbial markers across populations in early detection of colorectal cancer . Nat. Commun. 12 :3063. doi: 10.1038/s41467-021-23265-y, PMID: [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Xiao L., Zhang F., Zhao F. (2022). Large-scale microbiome data integration enables robust biomarker identification . Nat. Comput. Sci. 2 , 307–316. doi: 10.1038/s43588-022-00247-8 [ CrossRef ] [ Google Scholar ]
- Yeung A. W. K. (2019). Comparison between Scopus, web of science, pub med and publishers for mislabelled review papers . Curr. Sci. 116 :1909. doi: 10.18520/cs/v116/i11/1909-1914 [ CrossRef ] [ Google Scholar ]
- Yu D., Kou G., Xu Z., Shi S. (2020a). Analysis of collaboration evolution in AHP research: 1982–2018 . Int. J. Inf. Tech. Decis. 20 , 7–36. doi: 10.1142/S0219622020500406 [ CrossRef ] [ Google Scholar ]
- Yu D., Xu Z., Pedrycz W. (2020b). Bibliometric analysis of rough sets research . Appl. Soft Comput. 94 :106467. doi: 10.1016/j.asoc.2020.106467 [ CrossRef ] [ Google Scholar ]
- Yu D., Xu Z., Wang W. (2018). Bibliometric analysis of fuzzy theory research in China: a 30-year perspective . Knowl.-Based Syst. 141 , 188–199. doi: 10.1016/j.knosys.2017.11.018 [ CrossRef ] [ Google Scholar ]
- Zhang X., Li L., Butcher J., Stintzi A., Figeys D. (2019). Advancing functional and translational microbiome research using meta-omics approaches . Microbiome 7 :154. doi: 10.1186/s40168-019-0767-6, PMID: [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Zhao L., Xiong Q., Stary C. M., Mahgoub O. K., Ye Y., Gu L., et al.. (2018). Bidirectional gut-brain-microbiota axis as a potential link between inflammatory bowel disease and ischemic stroke . J. Neuroinflammation 15 :339. doi: 10.1186/s12974-018-1382-3, PMID: [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Zhou W., Sailani M. R., Contrepois K., Zhou Y., Ahadi S., Leopold S. R., et al.. (2019). Longitudinal multi-omics of host–microbe dynamics in prediabetes . Nature 569 , 663–671. doi: 10.1038/s41586-019-1236-x, PMID: [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Zhou Y., Xu Z. Z., He Y., Yang Y., Liu L., Lin Q., et al.. (2018). Gut microbiota offers universal biomarkers across ethnicity in inflammatory bowel disease diagnosis and infliximab response prediction . mSystems 3 , e00188–e00117. doi: 10.1128/mSystems.00188-17, PMID: [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
Supported by
Microbiology

Life After Asteroid Bennu
Dante Lauretta, the planetary scientist who led the OSIRIS-REx mission to retrieve a handful of space dust, discusses his next final frontier.
By Katrina Miller

Toddlers Smell Like Flowers, Teens Smell ‘Goatlike,’ Study Finds
Two musky steroids, and higher levels of odorous acids, distinguish the body odors of adolescents and tots.
By Emily Anthes

2 Scientists in Canada Passed On Secrets to China, Investigations Find
After a prolonged parliamentary debate, details about two microbiology researchers who were found to have shared secrets with China have been released.
By Ian Austen

It’s Probably Time to Clean Your Water Bottle
Yes, that black stuff is mold.
By Knvul Sheikh

Why Are California’s Waters Lighting Up in Blue?
It looks like the trailer for “Avatar: The Way of Water,” but it’s a natural phenomenon. And spotting it is getting increasingly popular.
By Claire Moses

Cómo saber si en realidad necesitas ese antibiótico
Se calcula que el 28 por ciento de los antibióticos recetados a niños y adultos son innecesarios. En esta temporada, aprende cómo usarlos (y cuándo es mejor evitarlos).
By Dawn MacKeen
Do You Really Need That Antibiotic?
It’s antibiotic season. Brush up on how you should use them — and when to avoid them.

Mars Needs Insects
If humans are ever going to live on the red planet, they’re going to have to bring bugs with them.
By Sarah Scoles

Male-Killing Virus Is Discovered in Insects
The chance finding in a Japanese university’s greenhouse could help researchers find ways to control agricultural pests or even insects that spread disease.
By Elizabeth Anne Brown

Old Faithful Is Boiling, Smelly and the Perfect Home for These Living Things
In one of Yellowstone National Park’s most well-known attractions, researchers discovered an extraordinary diversity of microbial life.
By Sarah Derouin
Advertisement

Microbiology
- Publishes experimental and theoretical articles, critical reviews, and short communications.
- The target audience is specialists at research institutions and medical workers.
- The journal welcomes manuscripts from all countries.
- Nikolai.V Pimenov

Latest issue
Volume 93, Issue 1
Latest articles
Green approach: characterization, antibacterial, antitumor and detoxification potentials of exopolysaccharides-mediated silver nanoparticles produced by penicillium citrinum aumc 11627.
- M. M. Housseiny
- E. M. Fawzy
- O. M. El-Mahdy

Immobilization of Azospirillum Bacteria on Various Carriers
- M. A. Kupryashina
- E. G. Ponomareva
- T. E. Pylaev

Characteristics of Psychrotolerant Pseudomonads Isolated from Organogenic Clay Deposits of the Mramornaya Cave (Primorskii Krai)
- D. A. Rusakova
- M. L. Sidorenko

Ecological Diversity of Micromycetes in Aerial Environments of Russian Libraries
- T. D. Velikova
- E. A. Popikhina
- S. S. Khazova

Chitin Degradation by Microbial Communities of the Kandalaksha Bay, White Sea
- A. M. Dukat
- A. M. Kuznetsova
- I. V. Danilova
Journal information
- Biological Abstracts
- Chemical Abstracts Service (CAS)
- Current Contents/Life Sciences
- Google Scholar
- Japanese Science and Technology Agency (JST)
- Norwegian Register for Scientific Journals and Series
- OCLC WorldCat Discovery Service
- Science Citation Index Expanded (SCIE)
- TD Net Discovery Service
- UGC-CARE List (India)
Rights and permissions
Springer policies
© Pleiades Publishing, Ltd.
- Find a journal
- Publish with us
- Track your research
- Search Menu
- FEMS Microbiology Ecology
- FEMS Microbiology Letters
- FEMS Microbiology Reviews
- FEMS Yeast Research
- Pathogens and Disease
- FEMS Microbes
- Awards & Prizes
- Editor's Choice Articles
- Thematic Issues
- Virtual Special Issues
- Call for Papers
- Journal Policies
- Open Access Options
- Submit to the FEMS Journals
- Why Publish with the FEMS Journals
- About the Federation of European Microbiological Societies
- About the FEMS Journals
- Advertising and Corporate Services
- Conference Reports
- Editorial Boards
- Investing in Science
- Journals Career Network
- Journals on Oxford Academic
- Books on Oxford Academic

Six Key Topics in Microbiology: 2024
This collection from the FEMS journals presents the latest high-quality research in six key topic areas of microbiology that have an impact across the world. All of the FEMS journals aim to serve the microbiology community with timely and authoritative research and reviews, and by investing back into the science community .
Interested in publishing your research relevant to the six key microbiology topics?
Learn more about why the FEMS journals are the perfect home for your microbiology research.
Browse the collection categories:
Antimicrobial resistance, environmental microbiology, pathogenicity and virulence, biotechnology and synthetic biology, microbiomes, food microbiology.

FEMS and Open Access: Embracing an Open Future
As of January 2024, FEMS has flipped four of its journals to fully open access (OA), making six out of its seven journals OA. FEMS Microbiology Letters remains a subscription journal and free to publish in.
We are excited to be making high quality science freely available to anyone to read anywhere in the world and further supporting the advancement of our discipline.
View our FAQs page

Never miss the latest research from the FEMS Journals
Stay up to date on the latest microbiology research with content alerts delivered directly to your inbox. This free service from OUP allows you to create custom email alerts to make sure you never miss our on the latest research from your favorite FEMS journals.
Learn more & sign up
Latest posts on X
Affiliations.
- Copyright © 2024
- About Oxford Academic
- Publish journals with us
- University press partners
- What we publish
- New features
- Open access
- Institutional account management
- Rights and permissions
- Get help with access
- Accessibility
- Advertising
- Media enquiries
- Oxford University Press
- Oxford Languages
- University of Oxford
Oxford University Press is a department of the University of Oxford. It furthers the University's objective of excellence in research, scholarship, and education by publishing worldwide
- Copyright © 2024 Oxford University Press
- Cookie settings
- Cookie policy
- Privacy policy
- Legal notice
This Feature Is Available To Subscribers Only
Sign In or Create an Account
This PDF is available to Subscribers Only
For full access to this pdf, sign in to an existing account, or purchase an annual subscription.

New enzymatic cocktail can kill tuberculosis-causing mycobacteria
W ith resistance to chemical antibiotics on the rise, the world needs entirely new forms of antibiotics. A new study published in Microbiology Spectrum shows that an enzymatic cocktail can kill a variety of mycobacterial species of bacteria, including those that cause tuberculosis. The research was carried out by scientists at Colorado State University and Endolytix Technologies.
"We have a mycobacterial drug that works for nontuberculous mycobacteria and M. tuberculosis that is biological, not phage therapy, and not small molecule antibiotics," said Jason Holder, Ph.D., a study co-author and Founder and Chief Science Officer at Endolytix Technology.
"Mycobacterial infections are particularly hard to treat due to poor efficacy with standard of care drugs that are used in multidrug regimens resulting in significant toxicities and treatments lasting six months to years. This is often followed up by reemergence of the bacterial infection after a year of testing negative."
In the new proof of principle study, the researchers took a biological approach instead of a chemical one to develop a cocktail of enzymes that attack the cell envelope of mycobacteria. The cocktail of enzymes contains highly specific biochemical catalysts that target and degrade the mycobacteria cell envelope that is essential for mycobacterial viability.
To increase efficacy, the researchers delivered the enzymatic drug inside of host macrophages where mycobacteria grow. In laboratory experiments, the drug was effective against M. tuberculosis and nontuberculous mycobacteria (NTMs), both lethal pulmonary lung diseases (PD).
TB kills roughly 1.5 million people per year.
"We characterized the mechanism of bactericide as through shredding of the bacterial cells into fragments," Holder said. "We've shown we can design and develop biological antibiotics and deliver them to the sites of infection through liposomal encapsulation. By combining drug delivery science with enzymes that lyse bacteria, we hope to open up treatment options in diseases such as NTM pulmonary disease, tuberculosis pulmonary disease and others."
According to study co-author Richard Slayden, Ph.D., a professor in the Department of Microbiology, Immunology and Pathology at Colorado State University, the new therapy complements current standard-of-care drugs and does not have many of the drug-drug interactions that are problematic with many anti-mycobacterial drugs in use.
"Endolytix enzymes work powerfully with standard-of-care antibiotics to kill bacteria with lower drug concentrations," Holder said. "This has the potential to reduce the significant toxicities associated with multi-drug regimens that are the standard for mycobacterial infections and hopefully lead to more rapid cures."
More information: Microbiology Spectrum (2024). journals.asm.org/doi/10.1128/spectrum.03534-23
Provided by American Society for Microbiology
- UB Directory

- Campaign >
- Find Your Cause >
- Featured Causes >
- Helping Students Now >
UB Awards 314 Biomedical Science Degrees; 18 Earn PhDs
Commencement 2023.

A graduate pays homage to her parents with words translated from Spanish meaning “For my parents, who arrived with nothing and gave me everything” on her mortarboard.
By Bill Bruton
Published June 1, 2023
Eighteen doctoral, 70 master’s and 226 baccalaureate candidates were eligible to receive degrees in biomedical science fields during the May commencement ceremony.
2023 Commencement Video

Related Links
- Commencement Program
- Full Gallery of Biomedical Sciences Commencement Photos
Three graduate students and 10 senior undergraduates were singled out for special honors, including two graduates who received a Chancellor’s Award, the highest State University of New York undergraduate honor.
Graduates completed work in 14 departments or programs of the Jacobs School of Medicine and Biomedical Sciences :
- biochemistry
- biomedical engineering
- biomedical informatics
- biomedical sciences
- biotechnical and clinical laboratory sciences
- genetics, genomics and bioinformatics
- medical physics
- microbiology and immunology
- natural sciences interdisciplinary
- neuroscience
- nuclear medicine technology
- pathology and anatomical sciences
- pharmacology and toxicology
- physiology and biophysics

Allison Brashear, MD, MBA, UB’s vice president for health sciences and dean of the Jacobs School, congratulates the Class of 2023.
Graduates also completed the following programs offered in alliance with the Roswell Park Comprehensive Cancer Center Graduate Division : cancer prevention and control, cancer sciences, cell and molecular biology and molecular pharmacology and cancer therapeutics.
Allison Brashear, MD, MBA , UB’s vice president for health sciences and dean of the Jacobs School, congratulated the graduates for their achievements.
“Despite hardships that all of us have faced the last few years, you have shown great resilience, determination and perseverance in your academic pursuits, qualities I am certain will enable you to make your mark in your respective industries,” Brashear said.
“You are the next generation of leaders, professionals, scientists and researchers,” Brashear added. “Your work will positively impact research aimed at ensuring equitable health outcomes across diverse patients and populations, while supporting the Jacobs School’s ongoing commitment to justice, equity, diversity and inclusion.”
Brashear emphasized the importance collaboration has had on their studies, and will continue to have in their careers.
“While your training may differ, at UB you learned that teamwork is essential, and that a diverse group working together can leverage its strengths and expertise to institute change,” Brashear said. “It is that type of collaboration that is fueling medical breakthroughs and the faster development of treatments and medications that impact patients’ lives.”
She spoke about how medical innovations — including artificial intelligence — will transform health care.
“This is only the beginning. The coming years will usher in a revolution in patient-centered care with the digitization of medical records, the development of biometric technology, and advancements in biology and life sciences,“ Brashear said. “While the industry is changing at a record speed, your UB degree has prepared you for a diverse job market and to remain versatile as scientists.”
“Your creative and critical thinking will continually push the bounds of scientific discovery and new technologies. It has been my honor to watch you grow and thrive, make new discoveries, and shape your goals to change the world,” Brashear added. “I applaud each and every one of you on achieving this next step in your journey and I wish you all the best in your bright futures.”
A. Scott Weber, PhD, provost and executive vice president for academic affairs, conferred the degrees during the May 21 event at UB’s Center for the Arts.
“Your UB education has prepared you to be flexible and responsive to the shifting needs and opportunities, and to meet the challenges we face head on, however vast they may seem,” Weber said.
“You’ve impacted our local and global communities through enhanced learning and engagement opportunities. Throughout your time at UB, you have proven that you are talented, compassionate, dedicated and strong. These truly are the attributes that will enable your future success,” Weber added.
Outstanding Graduates Recognized
Biochemistry graduate student research achievement award.
Doctoral graduate Christopher Campomizzi was honored for research that received national or international recognition and for being selected to give an oral presentation at a major national or international meeting.
Dissertation: “19F NMR Studies of CYP121 from Mycobacterium Tuberculosis Illustrate the Importance of the Protein Dimer”
Mentor: D. Fernando Estrada, PhD , associate professor of biochemistry
Roswell Park Graduate Division Award for Excellence in Research
Doctoral graduate Sarah Rose Chamberlain was the recipient of this award for outstanding research for her dissertation titled “Novel Approaches of Photodynamic Therapy for Lung Cancer”
Mentor: Gal Shafirstein, DSc , professor of oncology, Roswell Park Comprehensive Cancer Center
The Dean’s Award for Outstanding Dissertation Research
Doctoral graduate Murat Can Kalem was the winner of this award that recognizes demonstrated excellence in research.
He was honored for his dissertation: “Arginine Methylation and the Control of RNA-Binding Proteins in Cryptococcus neoformans”
Mentor: John C. Panepinto, PhD , professor of microbiology and immunology
The Microbiology and Immunology Award for Excellence in Dissertation Research in Memory of Dr. Murray W. Stinson
Doctoral graduate Murat Can Kalem was honored for his dissertation “Arginine Methylation and the Control of RNA-Binding Proteins in Cryptococcus neoformans”
SUNY Chancellor’s Award for Student Excellence
Haeni Lee and Richard Pasternack were recognized with the Chancellor’s Award. It recognizes students for their integration of academic excellence with other aspects of their lives that may include leadership, athletics, community service, creative and performing arts, entrepreneurship or career achievement.
Lee graduates with a Bachelor of Science degree in biomedical sciences with a public health minor. Lee is an international student from South Korea and a University Honors College Scholar.
She has been a senator in the Residence Hall Association, teaching assistant, research assistant and intern in several laboratories and a company. She has worked in the UB Maternal and Child Health program, contributing to work on helping pregnant women quit smoking and studying maternal substance use related to pandemic.
Pasternak, a native of Alden, New York, graduates with a Bachelor of Science degree in biomedical sciences. He is a recipient of UB’s Excellence in General and Organic Chemistry Awards and a member of Phi Beta Kappa.
Pasternack works as a research aide studying bicarbonate transport and is a medical assistant for an endocrinology practice. He volunteers at the Pediatric and Adolescent Urgent Care of WNY and is a board member for the Alden Community Scholarship Foundation.
Undergraduate Outstanding Senior Awards
The following awards honor high academic performance and involvement in the campus community and external organizations:
Biochemistry Kevin Bowman
Bioinformatics and Computational Biology, Biomedical Informatics Luna Liu
Biomedical Sciences Lily Freeman-Striegel
Biotechnology Fredrick Earl
Medical Technology Faith Dwyer
Neuroscience Colin Schupbach
Nuclear Medicine Technology Aja Holland
Pharmacology and Toxicology Jordan Richardson

Commencement speaker Arturo Casadevall, MD, PhD, tells the graduates they will play important roles in their research careers.
Infectious Disease Specialist is Speaker
Commencement speaker Arturo Casadevall, MD, PhD, distinguished professor and chair of molecular microbiology and immunology at Johns Hopkins University, told the graduates they will play an important role in shaping the world.
“I have a very simple message for you. You are humanity’s best insurance policy, and that makes you the most important scientific generation in the history of science,” Casadevall said. “This is no hyperbole, because you are graduating at a time of great challenges to our species.”
He went on to explain that when he got his degree, DNA sequencing was still a novelty, the life expectancy of someone with AIDS was a matter of months, the cause of cervical cancer was unknown, organ transplantation was rare and carried huge risks of organ rejection and infection, while ulcers were attributed to acid secretion and a type A personality.
Today, he said, DNA sequencing is commonplace, you can know your genetic ancestry for a few dollars, HIV is a treatable disease, there is a vaccine to prevent cervical cancer, organ transplantation is a routine procedure, and ulcers were found to be caused by bacteria and can now be treated with antibiotics.
“This progress was made possible because of new scientific knowledge. Basic science combined with clinical research to make discoveries that translated into new therapies,” Casadevall said. “To get here we needed new knowledge that in turn required approaching problems with the tools of science.”
The speaker also related that when he was 19, his father didn’t think he was going anywhere in life and insisted that Casadevall go to school to get a pest control operator license.
The classes were held at night in a community college in Brooklyn.
He still has that diploma proudly displayed in his office.
“Why am I telling you this? Because I want to make the point that life has many branch points and that the road to this podium was by no means straight or assured. In fact, I feel very lucky to have gotten as far as I have,” Casadevall said. “In medicine, I specialized in infectious disease and my research is in killing microbes. Hence, I am indeed in the business of killing bugs, and you could argue that I did take my father’s advice but just kept going.”
He offered hope to the graduates as they continue on in their research.
“I believe a day will come when one of you will give a commencement address, and you will tell the graduates that you remember a terrible time when many cancers were incurable, when many elderly individuals developed dementia, and when we feared that climate change was irreversible,” Casadevall said. “When that day comes, I hope you will reinforce the message that the way forward is to continue to generate knowledge to ensure an even better world built with the tools of science and ethics, on the wings of curiosity and the human spirit.”

IMAGES
VIDEO
COMMENTS
Microbiology is the study of microscopic organisms, such as bacteria, viruses, archaea, fungi and protozoa. This discipline includes fundamental research on the biochemistry, physiology, cell ...
Microbiology News. Articles and images on biochemistry research, micro-organisms, cell functions and related topics, updated daily.
To investigate the differences in bacterial and fungal community structure and diversity in conjunctival tissue of healthy and diabetic mice. Fengjiao Li, Shuo Yang, Ji Ma, Xiaowen Zhao, Meng Chen and Ye Wang. BMC Microbiology 2024 24 :90. Research Published on: 16 March 2024.
First published: January 18, 2024. Antimicrobial resistance (AMR) is a major global health issue. Current measures for tackling it comprise mainly the prudent use of drugs, the development of new drugs, and rapid diagnostics. Relatively little attention has been given to forecasting the evolution of resistance.
A recent study finds varying combinations of microbes in the vaginal microbiome may influence health outcomes such as risk of sexually transmitted disease and preterm birth. Microbiology coverage ...
As microbiology collaborations increased among researchers from different department and labs, Neil Rasmussen, a longtime member of the MIT Corporation and a member of the visiting committees for a number of departments, realized there was still one more component needed to turn microbiome research into a force for human health.
Current Research in Microbial Sciences publishes original papers and short communications, review articles as well as graphical reviews that cover all aspects of the broad field of microbiology, the study of microorganisms, or microbes, a diverse group of unicellular organisms and subcellular …. View full aims & scope. $1125.
Funding new ideas. Supported by Harvard Catalyst microbiome pilot funding over the past year through the Translational Innovator program, Gerber, Bry and Allegretti have been investigating whether a microbiome-focused clinical test can predict whose C. difficile infections will recur, which happens about 25 percent of the time.
The Emerging Roles of Microbiota-derived extracellular vesicles in psychiatric disorders. Chuang Guo. 白 玉龙. Pengfei Li. Kuanjun He. Frontiers in Microbiology. doi 10.3389/fmicb.2024.1383199. Original Research. Accepted on 28 Mar 2024.
The data of publications in the recent 5 years is used to assess the current research status of gut microbiome and disease. The timeline view of keywords co-occurrence network reveals the development of gut microbiome and disease (Supplementary Figure 10). This network is divided into 21 clusters, which present the major subtopics in this field.
In one of Yellowstone National Park's most well-known attractions, researchers discovered an extraordinary diversity of microbial life. News about Microbiology, including commentary and archival ...
Microbiology is now fully Open Access (OA). Find out more about the transformation of the Society's founding journal and its OA future here.As the founding journal from the Microbiology Society, Microbiology brings together communities of scientists from all microbiological disciplines and from around the world. Originally Journal of General Microbiology, we have been publishing the latest ...
Managing the rise in STIs among older adults. A new research review presented at a pre-congress day for this year's European Congress of Clinical Microbiology and Infectious Diseases (ECCMID 2024 ...
Molecular Pathogenesis of Enteroviruses: Insights into Viral-Host Interactions, Pathogenic Mechanisms, and Microbiome Dynamics. Man Lung Yeung. Jade LL Teng. Shubhada Bopegamage. 162 views. The most cited microbiology journal which advances our understanding of the role microbes play in addressing global challenges such as healthcare, food ...
Papers published in the journal cover all aspects of microbial taxonomy, phylogeny, ecology, physiology and metabolism, molecular genetics and genomics, gene regulation, viruses of prokaryotes, as well as interactions between …. View full aims & scope. Find out more about the IP. $2800. Article publishing charge.
Overview. Microbiology is an international peer-reviewed journal that addresses a broad spectrum of topics in both fundamental and applied microbiology. Publishes experimental and theoretical articles, critical reviews, and short communications. The target audience is specialists at research institutions and medical workers.
Six Key Topics in Microbiology: 2024. in Virtual Special Issues. This collection from the FEMS journals presents the latest high-quality research in six key topic areas of microbiology that have an impact across the world. All of the FEMS journals aim to serve the microbiology community with timely and authoritative research and reviews, and by ...
A new study published in Microbiology Spectrum shows that an enzymatic cocktail can kill a variety of mycobacterial species of bacteria, including those that cause tuberculosis. The research was ...
Pasternak, a native of Alden, New York, graduates with a Bachelor of Science degree in biomedical sciences. He is a recipient of UB's Excellence in General and Organic Chemistry Awards and a member of Phi Beta Kappa. Pasternack works as a research aide studying bicarbonate transport and is a medical assistant for an endocrinology practice.