When you choose to publish with PLOS, your research makes an impact. Make your work accessible to all, without restrictions, and accelerate scientific discovery with options like preprints and published peer review that make your work more Open.
- PLOS Biology
- PLOS Climate
- PLOS Complex Systems
- PLOS Computational Biology
- PLOS Digital Health
- PLOS Genetics
- PLOS Global Public Health
- PLOS Medicine
- PLOS Mental Health
- PLOS Neglected Tropical Diseases
- PLOS Pathogens
- PLOS Sustainability and Transformation
- PLOS Collections

Welcome to the PLOS Writing Center
Your source for scientific writing & publishing essentials.
A collection of free, practical guides and hands-on resources for authors looking to improve their scientific publishing skillset.
ARTICLE-WRITING ESSENTIALS
Your title is the first thing anyone who reads your article is going to see, and for many it will be where they stop reading. Learn how to write a title that helps readers find your article, draws your audience in and sets the stage for your research!
The abstract is your chance to let your readers know what they can expect from your article. Learn how to write a clear, and concise abstract that will keep your audience reading.
A clear methods section impacts editorial evaluation and readers’ understanding, and is also the backbone of transparency and replicability. Learn what to include in your methods section, and how much detail is appropriate.
In many fields, a statistical analysis forms the heart of both the methods and results sections of a manuscript. Learn how to report statistical analyses, and what other context is important for publication success and future reproducibility.
The discussion section contains the results and outcomes of a study. An effective discussion informs readers what can be learned from your experiment and provides context for the results.
Ensuring your manuscript is well-written makes it easier for editors, reviewers and readers to understand your work. Avoiding language errors can help accelerate review and minimize delays in the publication of your research.
The PLOS Writing Toolbox
Delivered to your inbox every two weeks, the Writing Toolbox features practical advice and tools you can use to prepare a research manuscript for submission success and build your scientific writing skillset.
Discover how to navigate the peer review and publishing process, beyond writing your article.
The path to publication can be unsettling when you’re unsure what’s happening with your paper. Learn about staple journal workflows to see the detailed steps required for ensuring a rigorous and ethical publication.
Reputable journals screen for ethics at submission—and inability to pass ethics checks is one of the most common reasons for rejection. Unfortunately, once a study has begun, it’s often too late to secure the requisite ethical reviews and clearances. Learn how to prepare for publication success by ensuring your study meets all ethical requirements before work begins.
From preregistration, to preprints, to publication—learn how and when to share your study.
How you store your data matters. Even after you publish your article, your data needs to be accessible and useable for the long term so that other researchers can continue building on your work. Good data management practices make your data discoverable and easy to use, promote a strong foundation for reproducibility and increase your likelihood of citations.
You’ve just spent months completing your study, writing up the results and submitting to your top-choice journal. Now the feedback is in and it’s time to revise. Set out a clear plan for your response to keep yourself on-track and ensure edits don’t fall through the cracks.
There’s a lot to consider when deciding where to submit your work. Learn how to choose a journal that will help your study reach its audience, while reflecting your values as a researcher.
Are you actively preparing a submission for a PLOS journal? Select the relevant journal below for more detailed guidelines.
How to Write an Article
Share the lessons of the Writing Center in a live, interactive training.
Access tried-and-tested training modules, complete with slides and talking points, workshop activities, and more.
- The Scientist University
The Fundamentals of Academic Science Writing
Writing is an essential skill for scientists, and learning how to write effectively starts with good fundamentals and lots of practice..

Nathan Ni holds a PhD from Queens University. He is a science editor for The Scientist’s Creative Services Team who strives to better understand and communicate the relationships between health and disease.
View full profile.
Learn about our editorial policies.

Writing is a big part of being a scientist, whether in the form of manuscripts, grants, reports, protocols, presentations, or even emails. However, many people look at writing as separate from science—a scientist writes, but scientists are not regarded as writers. 1 This outdated assertion means that writing and communication has been historically marginalized when it comes to training and educating new scientists. In truth, being a professional writer is part of being a scientist . 1 In today’s hypercompetitive academic environment, scientists need to be as proficient with the pen as they are with the pipette in order to showcase their work.
Using the Active Voice
Stereotypical academic writing is rigid, dry, and mechanical, delivering prose that evokes memories of high school and undergraduate laboratory reports. The hallmark of this stereotype is passive voice overuse. In writing, the passive voice is when the action comes at the end of a clause—for example, “the book was opened”. In scientific writing, it is particularly prevalent when detailing methodologies and results. How many times have we seen something like “citric acid was added to the solution, resulting in a two-fold reduction in pH” rather than “adding citric acid to the solution reduced the pH two-fold”?
Scientists should write in the active voice as much as possible. However, the active voice tends to place much more onus on the writer’s perspective, something that scientists have historically been instructed to stay away from. For example, “we treated the cells with phenylephrine” places much more emphasis on the operator than “the cells were treated with phenylephrine.” Furthermore, pronoun usage in academic writing is traditionally discouraged, but it is much harder, especially for those with non-native English proficiency, to properly use active voice without them.
Things are changing though, and scientists are recognizing the importance of giving themselves credit. Many major journals, including Nature , Science , PLoS One , and PNAS allow pronouns in their manuscripts, and prominent style guides such as APA even recommend using first-person pronouns, as traditional third-person writing can be ambiguous. 2 It is vital that a manuscript clearly and definitively highlights and states what the authors specifically did that was so important or novel, in contrast to what was already known. A simple “we found…” statement in the abstract and the introduction goes a long way towards giving readers the hook that they need to read further.
Keeping Sentences Simple
Writing in the active voice also makes it easier to organize manuscripts and construct arguments. Active voice uses fewer words than passive voice to explain the same concept. It also introduces argument components sequentially—subject, claim, and then evidence—whereas passive voice introduces claim and evidence before the subject. Compare, for example, “T cell abundance did not differ between wildtype and mutant mice” versus “there was no difference between wildtype and mutant mice in terms of T cell abundance.” T cell abundance, as the measured parameter, is the most important part of the sentence, but it is only introduced at the very end of the latter example.
The sequential nature of active voice therefore makes it easier to not get bogged down in overloading the reader with clauses and adhering to a general principle of “one sentence, one concept (or idea, or argument).” Consider the following sentence:
Research on CysLT 2 R , expressed in humans in umbilical vein endothelial cells, macrophages, platelets, the cardiac Purkinje system, and coronary endothelial cells , had been hampered by a lack of selective pharmacological agents , the majority of work instead using the nonselective cysLT antagonist/partial agonist Bay-u9773 or genetic models of CysLT 2 R expression modulation) .
The core message of this sentence is that CysLT 2 R research is hampered by a lack of selective pharmacological agents, but that message is muddled by the presence of two other major pieces of information: where CysLT 2 R is expressed and what researchers used to study CysLT 2 R instead of selective pharmacological agents. Because this sentence contains three main pieces of information, it is better to break it up into three separate sentences for clarity.
In humans, CysLT 2 R is expressed in umbilical vein endothelial cells, macrophages, platelets, the cardiac Purkinje system, and coronary endothelial cells . CysLT 2 R research has been hampered by a lack of selective pharmacological agents . Instead, the majority of work investigating the receptor has used either the nonselective cysLT antagonist/partial agonist Bay-u9773 or genetic models of CysLT 2 R expression modulation.
The Right Way to Apply Jargon
There is another key advantage to organizing sentences in this simple manner: it lets scientists manage how jargon is introduced to the reader. Jargon—special words used within a specific field or on a specific topic—is necessary in scientific writing. It is critical for succinctly describing key elements and explaining key concepts. But too much jargon can make a manuscript unreadable, either because the reader does not understand the terminology or because they are bogged down in reading all of the definitions.
The key to using jargon is to make it as easy as possible for the audience. General guidelines instruct writers to define new terms only when they are first used. However, it is cumbersome for a reader to backtrack considerable distances in a manuscript to look up a definition. If a term is first introduced in the introduction but not mentioned again until the discussion, the writer should re-define the term in a more casual manner. For example: “PI3K can be reversibly inhibited by LY294002 and irreversibly inhibited by wortmannin” in the introduction, accompanied by “when we applied the PI3K inhibitor LY294002” for the discussion. This not only makes things easier for the reader, but it also re-emphasizes what the scientist did and the results they obtained.
Practice Makes Better
Finally, the most important fundamental for science writing is to not treat it like a chore or a nuisance. Just as a scientist optimizes a bench assay through repeated trial and error, combined with literature reviews on what steps others have implemented, a scientist should practice, nurture, and hone their writing skills through repeated drafting, editing, and consultation. Do not be afraid to write. Putting pen to paper can help organize one’s thoughts, expose next steps for exploration, or even highlight additional experiments required to patch knowledge or logic gaps in existing studies.
Looking for more information on scientific writing? Check out The Scientist’s TS SciComm section. Looking for some help putting together a manuscript, a figure, a poster, or anything else? The Scientist’s Scientific Services may have the professional help that you need.
- Schimel J. Writing Science: How to Write Papers That Get Cited And Proposals That Get Funded . Oxford University Press; 2012.
- First-person pronouns. American Psychological Association. Updated July 2022. Accessed March 2024. https://apastyle.apa.org/style-grammar-guidelines/grammar/first-person-pronouns
Related community Research Resources

How Cloud Labs and Remote Research Shape Science

Artificial Neural Networks: Learning by Doing

Peer Profile Program
WRITING A SCIENTIFIC RESEARCH ARTICLE | Format for the paper | Edit your paper! | Useful books | FORMAT FOR THE PAPER Scientific research articles provide a method for scientists to communicate with other scientists about the results of their research. A standard format is used for these articles, in which the author presents the research in an orderly, logical manner. This doesn't necessarily reflect the order in which you did or thought about the work. This format is: | Title | Authors | Introduction | Materials and Methods | Results (with Tables and Figures ) | Discussion | Acknowledgments | Literature Cited | TITLE Make your title specific enough to describe the contents of the paper, but not so technical that only specialists will understand. The title should be appropriate for the intended audience. The title usually describes the subject matter of the article: Effect of Smoking on Academic Performance" Sometimes a title that summarizes the results is more effective: Students Who Smoke Get Lower Grades" AUTHORS 1. The person who did the work and wrote the paper is generally listed as the first author of a research paper. 2. For published articles, other people who made substantial contributions to the work are also listed as authors. Ask your mentor's permission before including his/her name as co-author. ABSTRACT 1. An abstract, or summary, is published together with a research article, giving the reader a "preview" of what's to come. Such abstracts may also be published separately in bibliographical sources, such as Biologic al Abstracts. They allow other scientists to quickly scan the large scientific literature, and decide which articles they want to read in depth. The abstract should be a little less technical than the article itself; you don't want to dissuade your potent ial audience from reading your paper. 2. Your abstract should be one paragraph, of 100-250 words, which summarizes the purpose, methods, results and conclusions of the paper. 3. It is not easy to include all this information in just a few words. Start by writing a summary that includes whatever you think is important, and then gradually prune it down to size by removing unnecessary words, while still retaini ng the necessary concepts. 3. Don't use abbreviations or citations in the abstract. It should be able to stand alone without any footnotes. INTRODUCTION What question did you ask in your experiment? Why is it interesting? The introduction summarizes the relevant literature so that the reader will understand why you were interested in the question you asked. One to fo ur paragraphs should be enough. End with a sentence explaining the specific question you asked in this experiment. MATERIALS AND METHODS 1. How did you answer this question? There should be enough information here to allow another scientist to repeat your experiment. Look at other papers that have been published in your field to get some idea of what is included in this section. 2. If you had a complicated protocol, it may helpful to include a diagram, table or flowchart to explain the methods you used. 3. Do not put results in this section. You may, however, include preliminary results that were used to design the main experiment that you are reporting on. ("In a preliminary study, I observed the owls for one week, and found that 73 % of their locomotor activity occurred during the night, and so I conducted all subsequent experiments between 11 pm and 6 am.") 4. Mention relevant ethical considerations. If you used human subjects, did they consent to participate. If you used animals, what measures did you take to minimize pain? RESULTS 1. This is where you present the results you've gotten. Use graphs and tables if appropriate, but also summarize your main findings in the text. Do NOT discuss the results or speculate as to why something happened; t hat goes in th e Discussion. 2. You don't necessarily have to include all the data you've gotten during the semester. This isn't a diary. 3. Use appropriate methods of showing data. Don't try to manipulate the data to make it look like you did more than you actually did. "The drug cured 1/3 of the infected mice, another 1/3 were not affected, and the third mouse got away." TABLES AND GRAPHS 1. If you present your data in a table or graph, include a title describing what's in the table ("Enzyme activity at various temperatures", not "My results".) For graphs, you should also label the x and y axes. 2. Don't use a table or graph just to be "fancy". If you can summarize the information in one sentence, then a table or graph is not necessary. DISCUSSION 1. Highlight the most significant results, but don't just repeat what you've written in the Results section. How do these results relate to the original question? Do the data support your hypothesis? Are your results consistent with what other investigators have reported? If your results were unexpected, try to explain why. Is there another way to interpret your results? What further research would be necessary to answer the questions raised by your results? How do y our results fit into the big picture? 2. End with a one-sentence summary of your conclusion, emphasizing why it is relevant. ACKNOWLEDGMENTS This section is optional. You can thank those who either helped with the experiments, or made other important contributions, such as discussing the protocol, commenting on the manuscript, or buying you pizza. REFERENCES (LITERATURE CITED) There are several possible ways to organize this section. Here is one commonly used way: 1. In the text, cite the literature in the appropriate places: Scarlet (1990) thought that the gene was present only in yeast, but it has since been identified in the platypus (Indigo and Mauve, 1994) and wombat (Magenta, et al., 1995). 2. In the References section list citations in alphabetical order. Indigo, A. C., and Mauve, B. E. 1994. Queer place for qwerty: gene isolation from the platypus. Science 275, 1213-1214. Magenta, S. T., Sepia, X., and Turquoise, U. 1995. Wombat genetics. In: Widiculous Wombats, Violet, Q., ed. New York: Columbia University Press. p 123-145. Scarlet, S.L. 1990. Isolation of qwerty gene from S. cerevisae. Journal of Unusual Results 36, 26-31. EDIT YOUR PAPER!!! "In my writing, I average about ten pages a day. Unfortunately, they're all the same page." Michael Alley, The Craft of Scientific Writing A major part of any writing assignment consists of re-writing. Write accurately Scientific writing must be accurate. Although writing instructors may tell you not to use the same word twice in a sentence, it's okay for scientific writing, which must be accurate. (A student who tried not to repeat the word "hamster" produced this confusing sentence: "When I put the hamster in a cage with the other animals, the little mammals began to play.") Make sure you say what you mean. Instead of: The rats were injected with the drug. (sounds like a syringe was filled with drug and ground-up rats and both were injected together) Write: I injected the drug into the rat.
- Be careful with commonly confused words:
Temperature has an effect on the reaction. Temperature affects the reaction.
I used solutions in various concentrations. (The solutions were 5 mg/ml, 10 mg/ml, and 15 mg/ml) I used solutions in varying concentrations. (The concentrations I used changed; sometimes they were 5 mg/ml, other times they were 15 mg/ml.)
Less food (can't count numbers of food) Fewer animals (can count numbers of animals)
A large amount of food (can't count them) A large number of animals (can count them)
The erythrocytes, which are in the blood, contain hemoglobin. The erythrocytes that are in the blood contain hemoglobin. (Wrong. This sentence implies that there are erythrocytes elsewhere that don't contain hemoglobin.)
Write clearly
1. Write at a level that's appropriate for your audience.
"Like a pigeon, something to admire as long as it isn't over your head." Anonymous
2. Use the active voice. It's clearer and more concise than the passive voice.
Instead of: An increased appetite was manifested by the rats and an increase in body weight was measured. Write: The rats ate more and gained weight.
3. Use the first person.
Instead of: It is thought Write: I think
Instead of: The samples were analyzed Write: I analyzed the samples
4. Avoid dangling participles.
"After incubating at 30 degrees C, we examined the petri plates." (You must've been pretty warm in there.)
Write succinctly
1. Use verbs instead of abstract nouns
Instead of: take into consideration Write: consider
2. Use strong verbs instead of "to be"
Instead of: The enzyme was found to be the active agent in catalyzing... Write: The enzyme catalyzed...
3. Use short words.
Instead of: Write: possess have sufficient enough utilize use demonstrate show assistance help terminate end
4. Use concise terms.
Instead of: Write: prior to before due to the fact that because in a considerable number of cases often the vast majority of most during the time that when in close proximity to near it has long been known that I'm too lazy to look up the reference
5. Use short sentences. A sentence made of more than 40 words should probably be rewritten as two sentences.
"The conjunction 'and' commonly serves to indicate that the writer's mind still functions even when no signs of the phenomenon are noticeable." Rudolf Virchow, 1928
Check your grammar, spelling and punctuation
1. Use a spellchecker, but be aware that they don't catch all mistakes.
"When we consider the animal as a hole,..." Student's paper
2. Your spellchecker may not recognize scientific terms. For the correct spelling, try Biotech's Life Science Dictionary or one of the technical dictionaries on the reference shelf in the Biology or Health Sciences libraries.
3. Don't, use, unnecessary, commas.
4. Proofread carefully to see if you any words out.
USEFUL BOOKS
Victoria E. McMillan, Writing Papers in the Biological Sciences , Bedford Books, Boston, 1997 The best. On sale for about $18 at Labyrinth Books, 112th Street. On reserve in Biology Library
Jan A. Pechenik, A Short Guide to Writing About Biology , Boston: Little, Brown, 1987
Harrison W. Ambrose, III & Katharine Peckham Ambrose, A Handbook of Biological Investigation , 4th edition, Hunter Textbooks Inc, Winston-Salem, 1987 Particularly useful if you need to use statistics to analyze your data. Copy on Reference shelf in Biology Library.
Robert S. Day, How to Write and Publish a Scientific Paper , 4th edition, Oryx Press, Phoenix, 1994. Earlier editions also good. A bit more advanced, intended for those writing papers for publication. Fun to read. Several copies available in Columbia libraries.
William Strunk, Jr. and E. B. White, The Elements of Style , 3rd ed. Macmillan, New York, 1987. Several copies available in Columbia libraries. Strunk's first edition is available on-line.
Write Like a Scientist
A Guide to Scientific Communication
What is scientific writing ?
Scientific writing is a technical form of writing that is designed to communicate scientific information to other scientists. Depending on the specific scientific genre—a journal article, a scientific poster, or a research proposal, for example—some aspects of the writing may change, such as its purpose , audience , or organization . Many aspects of scientific writing, however, vary little across these writing genres. Important hallmarks of all scientific writing are summarized below. Genre-specific information is located here and under the “By Genre” tab at the top of the page.
What are some important hallmarks of professional scientific writing?
1. Its primary audience is other scientists. Because of its intended audience, student-oriented or general-audience details, definitions, and explanations — which are often necessary in lab manuals or reports — are not terribly useful. Explaining general-knowledge concepts or how routine procedures were performed actually tends to obstruct clarity, make the writing wordy, and detract from its professional tone.
2. It is concise and precise . A goal of scientific writing is to communicate scientific information clearly and concisely. Flowery, ambiguous, wordy, and redundant language run counter to the purpose of the writing.
3. It must be set within the context of other published work. Because science builds on and corrects itself over time, scientific writing must be situated in and reference the findings of previous work . This context serves variously as motivation for new work being proposed or the paper being written, as points of departure or congruence for new findings and interpretations, and as evidence of the authors’ knowledge and expertise in the field.
All of the information under “The Essentials” tab is intended to help you to build your knowledge and skills as a scientific writer regardless of the scientific discipline you are studying or the specific assignment you might be working on. In addition to discussions of audience and purpose , professional conventions like conciseness and specificity, and how to find and use literature references appropriately, we also provide guidelines for how to organize your writing and how to avoid some common mechanical errors .
If you’re new to this site or to professional scientific writing, we recommend navigating the sub-sections under “The Essentials” tab in the order they’re provided. Once you’ve covered these essentials, you might find information on genre- or discipline-specific writing useful.
An official website of the United States government
The .gov means it’s official. Federal government websites often end in .gov or .mil. Before sharing sensitive information, make sure you’re on a federal government site.
The site is secure. The https:// ensures that you are connecting to the official website and that any information you provide is encrypted and transmitted securely.
- Publications
- Account settings
Preview improvements coming to the PMC website in October 2024. Learn More or Try it out now .
- Advanced Search
- Journal List

A Guide to Writing a Scientific Paper: A Focus on High School Through Graduate Level Student Research
Renee a. hesselbach.
1 NIEHS Children's Environmental Health Sciences Core Center, University of Wisconsin—Milwaukee, Milwaukee, Wisconsin.
David H. Petering
2 Department of Chemistry and Biochemistry, University of Wisconsin—Milwaukee, Milwaukee, Wisconsin.
Craig A. Berg
3 Curriculum and Instruction, University of Wisconsin—Milwaukee, Milwaukee, Wisconsin.
Henry Tomasiewicz
Daniel weber.
This article presents a detailed guide for high school through graduate level instructors that leads students to write effective and well-organized scientific papers. Interesting research emerges from the ability to ask questions, define problems, design experiments, analyze and interpret data, and make critical connections. This process is incomplete, unless new results are communicated to others because science fundamentally requires peer review and criticism to validate or discard proposed new knowledge. Thus, a concise and clearly written research paper is a critical step in the scientific process and is important for young researchers as they are mastering how to express scientific concepts and understanding. Moreover, learning to write a research paper provides a tool to improve science literacy as indicated in the National Research Council's National Science Education Standards (1996), and A Framework for K–12 Science Education (2011), the underlying foundation for the Next Generation Science Standards currently being developed. Background information explains the importance of peer review and communicating results, along with details of each critical component, the Abstract, Introduction, Methods, Results , and Discussion . Specific steps essential to helping students write clear and coherent research papers that follow a logical format, use effective communication, and develop scientific inquiry are described.
Introduction
A key part of the scientific process is communication of original results to others so that one's discoveries are passed along to the scientific community and the public for awareness and scrutiny. 1 – 3 Communication to other scientists ensures that new findings become part of a growing body of publicly available knowledge that informs how we understand the world around us. 2 It is also what fuels further research as other scientists incorporate novel findings into their thinking and experiments.
Depending upon the researcher's position, intent, and needs, communication can take different forms. The gold standard is writing scientific papers that describe original research in such a way that other scientists will be able to repeat it or to use it as a basis for their studies. 1 For some, it is expected that such articles will be published in scientific journals after they have been peer reviewed and accepted for publication. Scientists must submit their articles for examination by other scientists familiar with the area of research, who decide whether the work was conducted properly and whether the results add to the knowledge base and are conveyed well enough to merit publication. 2 If a manuscript passes the scrutiny of peer-review, it has the potential to be published. 1 For others, such as for high school or undergraduate students, publishing a research paper may not be the ultimate goal. However, regardless of whether an article is to be submitted for publication, peer review is an important step in this process. For student researchers, writing a well-organized research paper is a key step in learning how to express understanding, make critical connections, summarize data, and effectively communicate results, which are important goals for improving science literacy of the National Research Council's National Science Education Standards, 4 and A Framework for K–12 Science Education, 5 and the Next Generation Science Standards 6 currently being developed and described in The NSTA Reader's Guide to A Framework for K–12 Science Education. 7 Table 1 depicts the key skills students should develop as part of the Science as Inquiry Content Standard. Table 2 illustrates the central goals of A Framework for K–12 Science Education Scientific and Engineering Practices Dimension.
Key Skills of the Science as Inquiry National Science Education Content Standard
National Research Council (1996).
Important Practices of A Framework for K–12 Science Education Scientific and Engineering Practices Dimension
National Research Council (2011).
Scientific papers based on experimentation typically include five predominant sections: Abstract, Introduction, Methods, Results, and Discussion . This structure is a widely accepted approach to writing a research paper, and has specific sections that parallel the scientific method. Following this structure allows the scientist to tell a clear, coherent story in a logical format, essential to effective communication. 1 , 2 In addition, using a standardized format allows the reader to find specific information quickly and easily. While readers may not have time to read the entire research paper, the predictable format allows them to focus on specific sections such as the Abstract , Introduction , and Discussion sections. Therefore, it is critical that information be placed in the appropriate and logical section of the report. 3
Guidelines for Writing a Primary Research Article
The Title sends an important message to the reader about the purpose of the paper. For example, Ethanol Effects on the Developing Zebrafish: Neurobehavior and Skeletal Morphogenesis 8 tells the reader key information about the content of the research paper. Also, an appropriate and descriptive title captures the attention of the reader. When composing the Title , students should include either the aim or conclusion of the research, the subject, and possibly the independent or dependent variables. Often, the title is created after the body of the article has been written, so that it accurately reflects the purpose and content of the article. 1 , 3
The Abstract provides a short, concise summary of the research described in the body of the article and should be able to stand alone. It provides readers with a quick overview that helps them decide whether the article may be interesting to read. Included in the Abstract are the purpose or primary objectives of the experiment and why they are important, a brief description of the methods and approach used, key findings and the significance of the results, and how this work is different from the work of others. It is important to note that the Abstract briefly explains the implications of the findings, but does not evaluate the conclusions. 1 , 3 Just as with the Title , this section needs to be written carefully and succinctly. Often this section is written last to ensure it accurately reflects the content of the paper. Generally, the optimal length of the Abstract is one paragraph between 200 and 300 words, and does not contain references or abbreviations.
All new research can be categorized by field (e.g., biology, chemistry, physics, geology) and by area within the field (e.g., biology: evolution, ecology, cell biology, anatomy, environmental health). Many areas already contain a large volume of published research. The role of the Introduction is to place the new research within the context of previous studies in the particular field and area, thereby introducing the audience to the research and motivating the audience to continue reading. 1
Usually, the writer begins by describing what is known in the area that directly relates to the subject of the article's research. Clearly, this must be done judiciously; usually there is not room to describe every bit of information that is known. Each statement needs one or more references from the scientific literature that supports its validity. Students must be reminded to cite all references to eliminate the risk of plagiarism. 2 Out of this context, the author then explains what is not known and, therefore, what the article's research seeks to find out. In doing so, the scientist provides the rationale for the research and further develops why this research is important. The final statement in the Introduction should be a clearly worded hypothesis or thesis statement, as well as a brief summary of the findings as they relate to the stated hypothesis. Keep in mind that the details of the experimental findings are presented in the Results section and are aimed at filling the void in our knowledge base that has been pointed out in the Introduction .
Materials and Methods
Research utilizes various accepted methods to obtain the results that are to be shared with others in the scientific community. The quality of the results, therefore, depends completely upon the quality of the methods that are employed and the care with which they are applied. The reader will refer to the Methods section: (a) to become confident that the experiments have been properly done, (b) as the guide for repeating the experiments, and (c) to learn how to do new methods.
It is particularly important to keep in mind item (b). Since science deals with the objective properties of the physical and biological world, it is a basic axiom that these properties are independent of the scientist who reported them. Everyone should be able to measure or observe the same properties within error, if they do the same experiment using the same materials and procedures. In science, one does the same experiment by exactly repeating the experiment that has been described in the Methods section. Therefore, someone can only repeat an experiment accurately if all the relevant details of the experimental methods are clearly described. 1 , 3
The following information is important to include under illustrative headings, and is generally presented in narrative form. A detailed list of all the materials used in the experiments and, if important, their source should be described. These include biological agents (e.g., zebrafish, brine shrimp), chemicals and their concentrations (e.g., 0.20 mg/mL nicotine), and physical equipment (e.g., four 10-gallon aquariums, one light timer, one 10-well falcon dish). The reader needs to know as much as necessary about each of the materials; however, it is important not to include extraneous information. For example, consider an experiment involving zebrafish. The type and characteristics of the zebrafish used must be clearly described so another scientist could accurately replicate the experiment, such as 4–6-month-old male and female zebrafish, the type of zebrafish used (e.g., Golden), and where they were obtained (e.g., the NIEHS Children's Environmental Health Sciences Core Center in the WATER Institute of the University of Wisconsin—Milwaukee). In addition to describing the physical set-up of the experiment, it may be helpful to include photographs or diagrams in the report to further illustrate the experimental design.
A thorough description of each procedure done in the reported experiment, and justification as to why a particular method was chosen to most effectively answer the research question should also be included. For example, if the scientist was using zebrafish to study developmental effects of nicotine, the reader needs to know details about how and when the zebrafish were exposed to the nicotine (e.g., maternal exposure, embryo injection of nicotine, exposure of developing embryo to nicotine in the water for a particular length of time during development), duration of the exposure (e.g., a certain concentration for 10 minutes at the two-cell stage, then the embryos were washed), how many were exposed, and why that method was chosen. The reader would also need to know the concentrations to which the zebrafish were exposed, how the scientist observed the effects of the chemical exposure (e.g., microscopic changes in structure, changes in swimming behavior), relevant safety and toxicity concerns, how outcomes were measured, and how the scientist determined whether the data/results were significantly different in experimental and unexposed control animals (statistical methods).
Students must take great care and effort to write a good Methods section because it is an essential component of the effective communication of scientific findings.
The Results section describes in detail the actual experiments that were undertaken in a clear and well-organized narrative. The information found in the Methods section serves as background for understanding these descriptions and does not need to be repeated. For each different experiment, the author may wish to provide a subtitle and, in addition, one or more introductory sentences that explains the reason for doing the experiment. In a sense, this information is an extension of the Introduction in that it makes the argument to the reader why it is important to do the experiment. The Introduction is more general; this text is more specific.
Once the reader understands the focus of the experiment, the writer should restate the hypothesis to be tested or the information sought in the experiment. For example, “Atrazine is routinely used as a crop pesticide. It is important to understand whether it affects organisms that are normally found in soil. We decided to use worms as a test organism because they are important members of the soil community. Because atrazine damages nerve cells, we hypothesized that exposure to atrazine will inhibit the ability of worms to do locomotor activities. In the first experiment, we tested the effect of the chemical on burrowing action.”
Then, the experiments to be done are described and the results entered. In reporting on experimental design, it is important to identify the dependent and independent variables clearly, as well as the controls. The results must be shown in a way that can be reproduced by the reader, but do not include more details than needed for an effective analysis. Generally, meaningful and significant data are gathered together into tables and figures that summarize relevant information, and appropriate statistical analyses are completed based on the data gathered. Besides presenting each of these data sources, the author also provides a written narrative of the contents of the figures and tables, as well as an analysis of the statistical significance. In the narrative, the writer also connects the results to the aims of the experiment as described above. Did the results support the initial hypothesis? Do they provide the information that was sought? Were there problems in the experiment that compromised the results? Be careful not to include an interpretation of the results; that is reserved for the Discussion section.
The writer then moves on to the next experiment. Again, the first paragraph is developed as above, except this experiment is seen in the context of the first experiment. In other words, a story is being developed. So, one commonly refers to the results of the first experiment as part of the basis for undertaking the second experiment. “In the first experiment we observed that atrazine altered burrowing activity. In order to understand how that might occur, we decided to study its impact on the basic biology of locomotion. Our hypothesis was that atrazine affected neuromuscular junctions. So, we did the following experiment..”
The Results section includes a focused critical analysis of each experiment undertaken. A hallmark of the scientist is a deep skepticism about results and conclusions. “Convince me! And then convince me again with even better experiments.” That is the constant challenge. Without this basic attitude of doubt and willingness to criticize one's own work, scientists do not get to the level of concern about experimental methods and results that is needed to ensure that the best experiments are being done and the most reproducible results are being acquired. Thus, it is important for students to state any limitations or weaknesses in their research approach and explain assumptions made upfront in this section so the validity of the research can be assessed.
The Discussion section is the where the author takes an overall view of the work presented in the article. First, the main results from the various experiments are gathered in one place to highlight the significant results so the reader can see how they fit together and successfully test the original hypotheses of the experiment. Logical connections and trends in the data are presented, as are discussions of error and other possible explanations for the findings, including an analysis of whether the experimental design was adequate. Remember, results should not be restated in the Discussion section, except insofar as it is absolutely necessary to make a point.
Second, the task is to help the reader link the present work with the larger body of knowledge that was portrayed in the Introduction . How do the results advance the field, and what are the implications? What does the research results mean? What is the relevance? 1 , 3
Lastly, the author may suggest further work that needs to be done based on the new knowledge gained from the research.
Supporting Documentation and Writing Skills
Tables and figures are included to support the content of the research paper. These provide the reader with a graphic display of information presented. Tables and figures must have illustrative and descriptive titles, legends, interval markers, and axis labels, as appropriate; should be numbered in the order that they appear in the report; and include explanations of any unusual abbreviations.
The final section of the scientific article is the Reference section. When citing sources, it is important to follow an accepted standardized format, such as CSE (Council of Science Editors), APA (American Psychological Association), MLA (Modern Language Association), or CMS (Chicago Manual of Style). References should be listed in alphabetical order and original authors cited. All sources cited in the text must be included in the Reference section. 1
When writing a scientific paper, the importance of writing concisely and accurately to clearly communicate the message should be emphasized to students. 1 – 3 Students should avoid slang and repetition, as well as abbreviations that may not be well known. 1 If an abbreviation must be used, identify the word with the abbreviation in parentheses the first time the term is used. Using appropriate and correct grammar and spelling throughout are essential elements of a well-written report. 1 , 3 Finally, when the article has been organized and formatted properly, students are encouraged to peer review to obtain constructive criticism and then to revise the manuscript appropriately. Good scientific writing, like any kind of writing, is a process that requires careful editing and revision. 1
A key dimension of NRC's A Framework for K–12 Science Education , Scientific and Engineering Practices, and the developing Next Generation Science Standards emphasizes the importance of students being able to ask questions, define problems, design experiments, analyze and interpret data, draw conclusions, and communicate results. 5 , 6 In the Science Education Partnership Award (SEPA) program at the University of Wisconsin—Milwaukee, we found the guidelines presented in this article useful for high school science students because this group of students (and probably most undergraduates) often lack in understanding of, and skills to develop and write, the various components of an effective scientific paper. Students routinely need to focus more on the data collected and analyze what the results indicated in relation to the research question/hypothesis, as well as develop a detailed discussion of what they learned. Consequently, teaching students how to effectively organize and write a research report is a critical component when engaging students in scientific inquiry.
Acknowledgments
This article was supported by a Science Education Partnership Award (SEPA) grant (Award Number R25RR026299) from the National Institute of Environmental Health Sciences of the National Institutes of Health. The SEPA program at the University of Wisconsin—Milwaukee is part of the Children's Environmental Health Sciences Core Center, Community Outreach and Education Core, funded by the National Institute of Environmental Health Sciences (Award Number P30ES004184). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or the National Institute of Environmental Health Sciences.
Disclosure Statement
No competing financial interests exist.

Oxford University Press's Academic Insights for the Thinking World

Scientific writing as a research skill

Scientific Papers Made Easy
- By Stuart West and Lindsay Turnbull
- February 2 nd 2024
Scientific papers are often hard to read, even for specialists that work in the area. This matters because potential readers will often give up and do something else instead. And that means the paper will have less impact.
The fact that many scientific papers are hard to read is surprising. Scientists want others to read their papers—they don’t try to make them difficult to get through! So why does this problem arise? And how can we fix it?
The curse of knowledge
One problem is that scientists are incredibly knowledgeable about every detail of their research: from the studies that inspired them, to their methods and results, to the implications of those results. This means that they’re about as far away as it’s possible to be from someone who is new to the topic, so often they’re the worst person in the world to write up their study.
This problem is so common that it has even been given a name in psychology literature: the ‘curse of knowledge’. The curse means that people tend to unwittingly assume that others have the necessary background to understand what they are saying. Put simply, it’s easy for a scientist to miss out crucial points or steps because they’ve forgotten how important those things are for understanding their work.
Another aspect of the ‘curse of knowledge’ is that scientists tend to write like scientists. They use jargon, technical abbreviations, and phrases that they would never use in everyday speech. This ‘science speak’ usually makes things harder, not easier, for potential readers. This is particularly true with readers of interdisciplinary research, or with readers who are new to the specific subject area, who are less likely to know the meaning behind the jargon.
So how can we fix the writing problems that come from science speak and the curse of knowledge?
Writing as a research skill
The first step is that we need to acknowledge that writing is a skill that needs to be learnt, just like any other aspect of scientific research. Indeed, good writing can require a much longer learning period than many familiar research techniques. Once you have learnt how to pipette, you can do it, but writing is something that you can keep improving throughout your career.
Writing can be learnt in multiple ways. Courses can be run, usually for undergraduate or graduate students. But learning to write needs practise and motivation, and these courses are often run before the students need to write up their own research, An alternative is guide
books, that provide advice and tips, that writers can read and apply as they go along, as they produce the different sections of their paper. But what exactly needs to be learnt?
The next step is to pause and imagine potential readers. A potential reader is likely to be time-limited, stressed, and easily bored. They have a million other things to do and will take any excuse to give up on reading your paper. They might be a PhD student trying to get to grips with their subject, or a professor who doesn’t really have time to read papers anymore.
They key point is that they don’t have to read your paper—it’s the writer’s job to make them want to. This leads to a fundamental principle of scientific writing: the reader must come first. It is the job of the writer to help the reader understand the content of their paper by making things as clear and straightforward as possible.
Guiding principles
Unfortunately, putting the reader first does not always come naturally, and can require a change of thinking on the part of the writer. Luckily there are a few general principles that help with this:
- Keep it Simple. Use simple clear writing to make it as easy as possible for the reader.
- Assume nothing. A paper is more likely to be hard to read because it assumed too much, rather than because it was dumbed down too much.
- Keep it to essentials. A more focused paper will better at both getting the major points across and keeping the attention of a time-stressed reader.
- Tell your story. Good scientific writing tells a story. It tells the reader why the topic you have chosen is important, what you found out, and why that matters.
The beauty is in the details
The above advice might still seem a bit vague, but it’s just an overview. In our recent book, Scientific Writing Made Easy, we build upon these guiding principles to provide a toolkit for writing the different parts of a scientific paper. We provide both a structure for each section, and detailed tips for how to fill that structure out. We make writing easier and less scary.
Our toolkit can be applied to different types of paper across the life, human, and natural sciences. While there are important differences, a lot of the same principles can be applied whether someone is writing up a laboratory experiment, a mathematical model, or an observational field study.
Learn more about Scientific Papers Made Easy with this review from the Stated Clearly YouTube channel.
Stuart West, Professor of Evolutionary Biology, Department of Biology, University of Oxford, UK. Lindsay Turnbull, Professor of Plant Ecology, Department of Biology, University of Oxford, UK.
- Earth & Life Sciences
- Science & Medicine
Our Privacy Policy sets out how Oxford University Press handles your personal information, and your rights to object to your personal information being used for marketing to you or being processed as part of our business activities.
We will only use your personal information to register you for OUPblog articles.
Or subscribe to articles in the subject area by email or RSS
Related posts:

Recent Comments
I am disappointed by this so-called article. I have long been interested in this field; it combines two interests of mine – science and language. Sad to say, what we have here is a free ad disguised as a review. If it had been written by independent reviewers, I would have been glad to hear of it. As it stands, this book is crossed off my TBR list. Too bad.
Sorry you thought that SB – it wasn’t trying to look at all like a ‘review’. The blog was a summary of what we think are some some key points about writing – which are then expanded in our book (where there is loads more space!). Cheers Stu
Comments are closed.

International Artificial Intelligence Conference
IAIC 2023: Computer Networks and IoT pp 263–274 Cite as
Element Extraction from Computer Science Academic Papers for AI Survey Writing
- Fan Luo 8 &
- Xinguo Yu 8
- Conference paper
- First Online: 03 April 2024
Part of the book series: Communications in Computer and Information Science ((CCIS,volume 2060))
With the exponential growth of research papers, text summarization tools have emerged. However, existing text summarization tools merely extract existing sentences or words based on their frequency and may not be particularly well-suited for papers. To address this gap, this study develops a model based on DistilBERT, primarily focusing on information extraction and dataset labeling and augmentation techniques. The model’s central objective is entity recognition, aiming to identify two specific entities from the full text of research papers. The model takes these critical segments of papers as input and aims to identify the research problems and content contained within them. In response to the limitations of existing datasets, this research augments a dataset with over 4000 full-text arXiv computer algorithm papers through manual annotations.
The developed model demonstrates exceptional performance on several evaluation metrics, including accuracy, precision, F1 score, and recall. For comparative experiments, we employed several baseline models based on BERT. These results demonstrate the effectiveness of the proposed model. As part of a comparative experiment, we trained our models using three different dataset training methods. Additionally, to evaluate our dataset’s quality and underline the importance of full-text data, we manually annotated a random selection of 4000 papers from the ARXIV Data dataset, extracting only their titles and abstracts. As a result, Our proposed model outperforms all the baseline models, achieving an accuracy of 0.823 and an F1 Score of 0.798 and models trained on the proposed full-text annotated dataset outperform those trained on other datasets.
- Information Extraction
- Dataset Labeling and Augmentation
- Automating Literature Review
- Text Mining
This is a preview of subscription content, log in via an institution .
Buying options
- Available as PDF
- Read on any device
- Instant download
- Own it forever
- Available as EPUB and PDF
- Compact, lightweight edition
- Dispatched in 3 to 5 business days
- Free shipping worldwide - see info
Tax calculation will be finalised at checkout
Purchases are for personal use only
Felizardo, K.R., Carver, J.C.: Automating systematic literature review. In: Contemporary Empirical Methods in Software Engineering, pp. 327–355. Springer, Cham (2020). https://doi.org/10.1007/978-3-030-32489-6_12
Chapter Google Scholar
McNabb, L., Laramee, R.S.: How to write a visualization survey paper: a starting point. In: Eurographics (Education Papers), pp. 29–39 (2019)
Google Scholar
Loza, V., Lahiri, S., Mihalcea, R., et al.: Building a dataset for summarization and keyword extraction from emails. In: LREC, pp. 2441–2446 (2014)
Jonnalagadda, S., Goyal, P., Huffman, M.: Automating data extraction in systematic reviews: a systematic review. Syst. Rev. 4 (1), 78 (2015)
Article Google Scholar
Aliyu, M.B., Iqbal, R., James, A.: The canonical model of structure for data extraction in systematic reviews of scientific research articles. In: 15th International Conference on Social Networks Analysis, Management and Security (SNAMS 2018), pp. 264–271 (2018)
Cabot, P.L.H., Navigli, R.: REBEL: relation extraction by end-to-end language generation. In: Findings of the Association for Computational Linguistics, EMNLP 2021, pp. 2370–2381 (2021)
Kenton, J.D.M.W.C., Toutanova, L.K.: BERT: pre-training of deep bidirectional transformers for language understanding. In: Proceedings of NAACL-HLT, vol. 1, p. 2 (2019)
Nayak, T., Ng, H.T.: Effective modeling of encoder-decoder architecture for joint entity and relation extraction. In: Proceedings of the AAAI Conference on Artificial Intelligence, vol. 34, no. 05, pp. 8528–8535 (2020)
Yamada, I., Asai, A., Shindo, H., et al.: LUKE: deep contextualized entity representations with entity-aware self-attention. arXiv preprint arXiv:2010.01057 (2020)
Zhang, R.H., Liu, Q., Fan, A.X., et al.: Minimize exposure bias of Seq2Seq models in joint entity and relation extraction. arXiv preprint arXiv:2009.07503 (2020)
Blloshmi, R., Conia, S., Tripodi, R., et al.: Generating senses and RoLes: an end-to-end model for dependency-and span-based semantic role labeling. In: IJCAI, pp. 3786–3793 (2021)
Dernoncourt, F., Lee, J.Y.: PubMed 200k RCT: a dataset for sequential sentence classification in medical abstracts. arXiv preprint arXiv:1710.06071 (2017)
Gehrke, J., Ginsparg, P., Kleinberg, J.: Overview of the 2003 KDD cup. ACM SIGKDD Explor. Newsl. 5 (2), 149–151 (2003)
Download references
Author information
Authors and affiliations.
Central China Normal University, Wuhan, China
Fan Luo & Xinguo Yu
You can also search for this author in PubMed Google Scholar
Corresponding author
Correspondence to Xinguo Yu .
Editor information
Editors and affiliations.
Huazhong University of Science and Technology, Wuhan, Hubei, China
Chinese Academy of Science, Shenzhen, China
Nanjing University of Science and Technology, Nanjing, China
Jianfeng Lu
Rights and permissions
Reprints and permissions
Copyright information
© 2024 The Author(s), under exclusive license to Springer Nature Singapore Pte Ltd.
About this paper
Cite this paper.
Luo, F., Yu, X. (2024). Element Extraction from Computer Science Academic Papers for AI Survey Writing. In: Jin, H., Pan, Y., Lu, J. (eds) Computer Networks and IoT. IAIC 2023. Communications in Computer and Information Science, vol 2060. Springer, Singapore. https://doi.org/10.1007/978-981-97-1332-5_21
Download citation
DOI : https://doi.org/10.1007/978-981-97-1332-5_21
Published : 03 April 2024
Publisher Name : Springer, Singapore
Print ISBN : 978-981-97-1331-8
Online ISBN : 978-981-97-1332-5
eBook Packages : Computer Science Computer Science (R0)
Share this paper
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Publish with us
Policies and ethics
- Find a journal
- Track your research

Research paper writing service 24/7: low price and fast result
Writing a paper in a modern world.
Even living in the 21st century, with no necessity to go to the libraries anymore, to collect the wisdom of teachers and philosophers, to travel across the globe, to make long notes to professors on letter writing paper and wait for their reply, it might be actually hard to compose a worthy narration anyway.
Yes, modern time is full of advantages! For example, technologies.
There is no more need to sit in front of blank paper to write on, people are using their PC and laptops to work anywhere they choose.
Planning to go to the park to get an inspiration? Love working in your favorite cafe? Do everything you like!
Remember Harry Potter and his necessity to visit libraries and carry heavy books with him? Staying up all night to correcting mistakes he made on his parchment writing paper? Luckily, you won’t face the same obstacles.
Availability of research paper writing services makes it easier for you to finish your project on time and still have an opportunity to enjoy your life (and not piling your table with books)! Let’s talk a bit about its advantages.
Why order online?
Remember good old times when teacher was giving out primary writing paper and all you have to do was to fill in the blanks and write three sentences and a drawing to make everyone happy? Well, life has become more complicated since then.
Or let’s dig deeper into the past! What about kindergarten, when children are given colorful fundations writing paper? We are more than sure that any professor (or even a high school) teacher won’t appreciate such a level of work. Research projects are way harder than all these childish games.
Research paper writing service is ready whenever you’re ready. That’s its main advantage. With 24/7 customer service there’s no need to worry about time zones or late hours. That ensures a quick process and helps you to write a paper without any worries about deadlines.
Happy clients will ensure you that this service is a life saver! And your part is easy here: just type “write my paper” in a search bar and enjoy!

CEE Comm Lab helps first-year undergraduates present scientific research
The following is a modified excerpt from the MIT News article, “ First-year MIT students gain hands-on research experience in supportive peer community ” by Callie Ayoub.
During MIT’s Independent Activities Period (IAP) this January, first-year students interested in civil and environmental engineering (CEE) participated in a four-week undergraduate research opportunities program known as the mini-UROP (1.097). The six-unit subject pairs first-year students with a CEE graduate student or postdoc mentor, providing them with an inside look at the research being conducted in the department. The program culminates with a presentation event open to the entire CEE community.
Overall, eight labs in the department opened their doors to the 2024 cohort, who were eager to take advantage of the opportunity to collaborate with current students and build a community around their interests. The interdisciplinary nature of the department’s research offered participants a wide range of projects to work on, from redefining autonomous vehicle deployment to mitigating the effects of drought on crops.

Mini-UROP participant Iraira Rivera Rojas works in the Marelli Lab in CEE.
Throughout the duration of the mini-UROP, participants attended three workshops led by Jared Berezin , the manager of the Civil and Environmental Engineering Communication Lab (CEE Comm Lab). The communication lab is a free resource to undergraduates, graduates, and postdocs in the CEE community, providing one-on-one coaching and interactive workshops. Held on Fridays during IAP, the workshops focused on visual and oral communication strategies to help students contextualize their projects, explain scientific concepts, describe their methodologies, and present their results.
“Students were fortunate to have research mentors in the lab, and my goal was to provide communication mentorship outside of the lab,” says Berezin. “Our weekly workshops focused on scientific communication strategies, but perhaps more importantly I’d prompt them to talk about their projects, ask questions, and brainstorm together. They really embraced the opportunity to foster a supportive peer community, which I think is a core part of the CEE experience.”

Mini-UROP participants present their research to fellow students, staff, and faculty.
A significant challenge students face while completing the program is condensing their research down to a clear and concise two-minute presentation. To assist with this task, the workshops also featured presentations by CEE Communication Fellows Ignacio Arzuaga and Matthew Goss , providing students with a preview of how their own presentations may take shape. Before the final presentation event, students also had the option to meet with Comm Fellows to receive feedback, rehearse their talks, and practice responding to questions about their work.
“The final talks were impressive, and I was proud of the students for approaching both their research and communication challenges with such curiosity and thoughtfulness,” Berezin remarks.
To learn more about the experiences of students and mentors during the 2024 mini-UROP, you can read the full MIT News article .
Thank you for visiting nature.com. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser (or turn off compatibility mode in Internet Explorer). In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript.
- View all journals
- My Account Login
- Explore content
- About the journal
- Publish with us
- Sign up for alerts
- Open access
- Published: 26 March 2024
Predicting and improving complex beer flavor through machine learning
- Michiel Schreurs ORCID: orcid.org/0000-0002-9449-5619 1 , 2 , 3 na1 ,
- Supinya Piampongsant 1 , 2 , 3 na1 ,
- Miguel Roncoroni ORCID: orcid.org/0000-0001-7461-1427 1 , 2 , 3 na1 ,
- Lloyd Cool ORCID: orcid.org/0000-0001-9936-3124 1 , 2 , 3 , 4 ,
- Beatriz Herrera-Malaver ORCID: orcid.org/0000-0002-5096-9974 1 , 2 , 3 ,
- Christophe Vanderaa ORCID: orcid.org/0000-0001-7443-5427 4 ,
- Florian A. Theßeling 1 , 2 , 3 ,
- Łukasz Kreft ORCID: orcid.org/0000-0001-7620-4657 5 ,
- Alexander Botzki ORCID: orcid.org/0000-0001-6691-4233 5 ,
- Philippe Malcorps 6 ,
- Luk Daenen 6 ,
- Tom Wenseleers ORCID: orcid.org/0000-0002-1434-861X 4 &
- Kevin J. Verstrepen ORCID: orcid.org/0000-0002-3077-6219 1 , 2 , 3
Nature Communications volume 15 , Article number: 2368 ( 2024 ) Cite this article
44k Accesses
797 Altmetric
Metrics details
- Chemical engineering
- Gas chromatography
- Machine learning
- Metabolomics
- Taste receptors
The perception and appreciation of food flavor depends on many interacting chemical compounds and external factors, and therefore proves challenging to understand and predict. Here, we combine extensive chemical and sensory analyses of 250 different beers to train machine learning models that allow predicting flavor and consumer appreciation. For each beer, we measure over 200 chemical properties, perform quantitative descriptive sensory analysis with a trained tasting panel and map data from over 180,000 consumer reviews to train 10 different machine learning models. The best-performing algorithm, Gradient Boosting, yields models that significantly outperform predictions based on conventional statistics and accurately predict complex food features and consumer appreciation from chemical profiles. Model dissection allows identifying specific and unexpected compounds as drivers of beer flavor and appreciation. Adding these compounds results in variants of commercial alcoholic and non-alcoholic beers with improved consumer appreciation. Together, our study reveals how big data and machine learning uncover complex links between food chemistry, flavor and consumer perception, and lays the foundation to develop novel, tailored foods with superior flavors.
Similar content being viewed by others
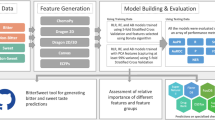
BitterSweet: Building machine learning models for predicting the bitter and sweet taste of small molecules
Rudraksh Tuwani, Somin Wadhwa & Ganesh Bagler
Sensory lexicon and aroma volatiles analysis of brewing malt
Xiaoxia Su, Miao Yu, … Tianyi Du
Predicting odor from molecular structure: a multi-label classification approach
Kushagra Saini & Venkatnarayan Ramanathan
Introduction
Predicting and understanding food perception and appreciation is one of the major challenges in food science. Accurate modeling of food flavor and appreciation could yield important opportunities for both producers and consumers, including quality control, product fingerprinting, counterfeit detection, spoilage detection, and the development of new products and product combinations (food pairing) 1 , 2 , 3 , 4 , 5 , 6 . Accurate models for flavor and consumer appreciation would contribute greatly to our scientific understanding of how humans perceive and appreciate flavor. Moreover, accurate predictive models would also facilitate and standardize existing food assessment methods and could supplement or replace assessments by trained and consumer tasting panels, which are variable, expensive and time-consuming 7 , 8 , 9 . Lastly, apart from providing objective, quantitative, accurate and contextual information that can help producers, models can also guide consumers in understanding their personal preferences 10 .
Despite the myriad of applications, predicting food flavor and appreciation from its chemical properties remains a largely elusive goal in sensory science, especially for complex food and beverages 11 , 12 . A key obstacle is the immense number of flavor-active chemicals underlying food flavor. Flavor compounds can vary widely in chemical structure and concentration, making them technically challenging and labor-intensive to quantify, even in the face of innovations in metabolomics, such as non-targeted metabolic fingerprinting 13 , 14 . Moreover, sensory analysis is perhaps even more complicated. Flavor perception is highly complex, resulting from hundreds of different molecules interacting at the physiochemical and sensorial level. Sensory perception is often non-linear, characterized by complex and concentration-dependent synergistic and antagonistic effects 15 , 16 , 17 , 18 , 19 , 20 , 21 that are further convoluted by the genetics, environment, culture and psychology of consumers 22 , 23 , 24 . Perceived flavor is therefore difficult to measure, with problems of sensitivity, accuracy, and reproducibility that can only be resolved by gathering sufficiently large datasets 25 . Trained tasting panels are considered the prime source of quality sensory data, but require meticulous training, are low throughput and high cost. Public databases containing consumer reviews of food products could provide a valuable alternative, especially for studying appreciation scores, which do not require formal training 25 . Public databases offer the advantage of amassing large amounts of data, increasing the statistical power to identify potential drivers of appreciation. However, public datasets suffer from biases, including a bias in the volunteers that contribute to the database, as well as confounding factors such as price, cult status and psychological conformity towards previous ratings of the product.
Classical multivariate statistics and machine learning methods have been used to predict flavor of specific compounds by, for example, linking structural properties of a compound to its potential biological activities or linking concentrations of specific compounds to sensory profiles 1 , 26 . Importantly, most previous studies focused on predicting organoleptic properties of single compounds (often based on their chemical structure) 27 , 28 , 29 , 30 , 31 , 32 , 33 , thus ignoring the fact that these compounds are present in a complex matrix in food or beverages and excluding complex interactions between compounds. Moreover, the classical statistics commonly used in sensory science 34 , 35 , 36 , 37 , 38 , 39 require a large sample size and sufficient variance amongst predictors to create accurate models. They are not fit for studying an extensive set of hundreds of interacting flavor compounds, since they are sensitive to outliers, have a high tendency to overfit and are less suited for non-linear and discontinuous relationships 40 .
In this study, we combine extensive chemical analyses and sensory data of a set of different commercial beers with machine learning approaches to develop models that predict taste, smell, mouthfeel and appreciation from compound concentrations. Beer is particularly suited to model the relationship between chemistry, flavor and appreciation. First, beer is a complex product, consisting of thousands of flavor compounds that partake in complex sensory interactions 41 , 42 , 43 . This chemical diversity arises from the raw materials (malt, yeast, hops, water and spices) and biochemical conversions during the brewing process (kilning, mashing, boiling, fermentation, maturation and aging) 44 , 45 . Second, the advent of the internet saw beer consumers embrace online review platforms, such as RateBeer (ZX Ventures, Anheuser-Busch InBev SA/NV) and BeerAdvocate (Next Glass, inc.). In this way, the beer community provides massive data sets of beer flavor and appreciation scores, creating extraordinarily large sensory databases to complement the analyses of our professional sensory panel. Specifically, we characterize over 200 chemical properties of 250 commercial beers, spread across 22 beer styles, and link these to the descriptive sensory profiling data of a 16-person in-house trained tasting panel and data acquired from over 180,000 public consumer reviews. These unique and extensive datasets enable us to train a suite of machine learning models to predict flavor and appreciation from a beer’s chemical profile. Dissection of the best-performing models allows us to pinpoint specific compounds as potential drivers of beer flavor and appreciation. Follow-up experiments confirm the importance of these compounds and ultimately allow us to significantly improve the flavor and appreciation of selected commercial beers. Together, our study represents a significant step towards understanding complex flavors and reinforces the value of machine learning to develop and refine complex foods. In this way, it represents a stepping stone for further computer-aided food engineering applications 46 .
To generate a comprehensive dataset on beer flavor, we selected 250 commercial Belgian beers across 22 different beer styles (Supplementary Fig. S1 ). Beers with ≤ 4.2% alcohol by volume (ABV) were classified as non-alcoholic and low-alcoholic. Blonds and Tripels constitute a significant portion of the dataset (12.4% and 11.2%, respectively) reflecting their presence on the Belgian beer market and the heterogeneity of beers within these styles. By contrast, lager beers are less diverse and dominated by a handful of brands. Rare styles such as Brut or Faro make up only a small fraction of the dataset (2% and 1%, respectively) because fewer of these beers are produced and because they are dominated by distinct characteristics in terms of flavor and chemical composition.
Extensive analysis identifies relationships between chemical compounds in beer
For each beer, we measured 226 different chemical properties, including common brewing parameters such as alcohol content, iso-alpha acids, pH, sugar concentration 47 , and over 200 flavor compounds (Methods, Supplementary Table S1 ). A large portion (37.2%) are terpenoids arising from hopping, responsible for herbal and fruity flavors 16 , 48 . A second major category are yeast metabolites, such as esters and alcohols, that result in fruity and solvent notes 48 , 49 , 50 . Other measured compounds are primarily derived from malt, or other microbes such as non- Saccharomyces yeasts and bacteria (‘wild flora’). Compounds that arise from spices or staling are labeled under ‘Others’. Five attributes (caloric value, total acids and total ester, hop aroma and sulfur compounds) are calculated from multiple individually measured compounds.
As a first step in identifying relationships between chemical properties, we determined correlations between the concentrations of the compounds (Fig. 1 , upper panel, Supplementary Data 1 and 2 , and Supplementary Fig. S2 . For the sake of clarity, only a subset of the measured compounds is shown in Fig. 1 ). Compounds of the same origin typically show a positive correlation, while absence of correlation hints at parameters varying independently. For example, the hop aroma compounds citronellol, and alpha-terpineol show moderate correlations with each other (Spearman’s rho=0.39 and 0.57), but not with the bittering hop component iso-alpha acids (Spearman’s rho=0.16 and −0.07). This illustrates how brewers can independently modify hop aroma and bitterness by selecting hop varieties and dosage time. If hops are added early in the boiling phase, chemical conversions increase bitterness while aromas evaporate, conversely, late addition of hops preserves aroma but limits bitterness 51 . Similarly, hop-derived iso-alpha acids show a strong anti-correlation with lactic acid and acetic acid, likely reflecting growth inhibition of lactic acid and acetic acid bacteria, or the consequent use of fewer hops in sour beer styles, such as West Flanders ales and Fruit beers, that rely on these bacteria for their distinct flavors 52 . Finally, yeast-derived esters (ethyl acetate, ethyl decanoate, ethyl hexanoate, ethyl octanoate) and alcohols (ethanol, isoamyl alcohol, isobutanol, and glycerol), correlate with Spearman coefficients above 0.5, suggesting that these secondary metabolites are correlated with the yeast genetic background and/or fermentation parameters and may be difficult to influence individually, although the choice of yeast strain may offer some control 53 .

Spearman rank correlations are shown. Descriptors are grouped according to their origin (malt (blue), hops (green), yeast (red), wild flora (yellow), Others (black)), and sensory aspect (aroma, taste, palate, and overall appreciation). Please note that for the chemical compounds, for the sake of clarity, only a subset of the total number of measured compounds is shown, with an emphasis on the key compounds for each source. For more details, see the main text and Methods section. Chemical data can be found in Supplementary Data 1 , correlations between all chemical compounds are depicted in Supplementary Fig. S2 and correlation values can be found in Supplementary Data 2 . See Supplementary Data 4 for sensory panel assessments and Supplementary Data 5 for correlation values between all sensory descriptors.
Interestingly, different beer styles show distinct patterns for some flavor compounds (Supplementary Fig. S3 ). These observations agree with expectations for key beer styles, and serve as a control for our measurements. For instance, Stouts generally show high values for color (darker), while hoppy beers contain elevated levels of iso-alpha acids, compounds associated with bitter hop taste. Acetic and lactic acid are not prevalent in most beers, with notable exceptions such as Kriek, Lambic, Faro, West Flanders ales and Flanders Old Brown, which use acid-producing bacteria ( Lactobacillus and Pediococcus ) or unconventional yeast ( Brettanomyces ) 54 , 55 . Glycerol, ethanol and esters show similar distributions across all beer styles, reflecting their common origin as products of yeast metabolism during fermentation 45 , 53 . Finally, low/no-alcohol beers contain low concentrations of glycerol and esters. This is in line with the production process for most of the low/no-alcohol beers in our dataset, which are produced through limiting fermentation or by stripping away alcohol via evaporation or dialysis, with both methods having the unintended side-effect of reducing the amount of flavor compounds in the final beer 56 , 57 .
Besides expected associations, our data also reveals less trivial associations between beer styles and specific parameters. For example, geraniol and citronellol, two monoterpenoids responsible for citrus, floral and rose flavors and characteristic of Citra hops, are found in relatively high amounts in Christmas, Saison, and Brett/co-fermented beers, where they may originate from terpenoid-rich spices such as coriander seeds instead of hops 58 .
Tasting panel assessments reveal sensorial relationships in beer
To assess the sensory profile of each beer, a trained tasting panel evaluated each of the 250 beers for 50 sensory attributes, including different hop, malt and yeast flavors, off-flavors and spices. Panelists used a tasting sheet (Supplementary Data 3 ) to score the different attributes. Panel consistency was evaluated by repeating 12 samples across different sessions and performing ANOVA. In 95% of cases no significant difference was found across sessions ( p > 0.05), indicating good panel consistency (Supplementary Table S2 ).
Aroma and taste perception reported by the trained panel are often linked (Fig. 1 , bottom left panel and Supplementary Data 4 and 5 ), with high correlations between hops aroma and taste (Spearman’s rho=0.83). Bitter taste was found to correlate with hop aroma and taste in general (Spearman’s rho=0.80 and 0.69), and particularly with “grassy” noble hops (Spearman’s rho=0.75). Barnyard flavor, most often associated with sour beers, is identified together with stale hops (Spearman’s rho=0.97) that are used in these beers. Lactic and acetic acid, which often co-occur, are correlated (Spearman’s rho=0.66). Interestingly, sweetness and bitterness are anti-correlated (Spearman’s rho = −0.48), confirming the hypothesis that they mask each other 59 , 60 . Beer body is highly correlated with alcohol (Spearman’s rho = 0.79), and overall appreciation is found to correlate with multiple aspects that describe beer mouthfeel (alcohol, carbonation; Spearman’s rho= 0.32, 0.39), as well as with hop and ester aroma intensity (Spearman’s rho=0.39 and 0.35).
Similar to the chemical analyses, sensorial analyses confirmed typical features of specific beer styles (Supplementary Fig. S4 ). For example, sour beers (Faro, Flanders Old Brown, Fruit beer, Kriek, Lambic, West Flanders ale) were rated acidic, with flavors of both acetic and lactic acid. Hoppy beers were found to be bitter and showed hop-associated aromas like citrus and tropical fruit. Malt taste is most detected among scotch, stout/porters, and strong ales, while low/no-alcohol beers, which often have a reputation for being ‘worty’ (reminiscent of unfermented, sweet malt extract) appear in the middle. Unsurprisingly, hop aromas are most strongly detected among hoppy beers. Like its chemical counterpart (Supplementary Fig. S3 ), acidity shows a right-skewed distribution, with the most acidic beers being Krieks, Lambics, and West Flanders ales.
Tasting panel assessments of specific flavors correlate with chemical composition
We find that the concentrations of several chemical compounds strongly correlate with specific aroma or taste, as evaluated by the tasting panel (Fig. 2 , Supplementary Fig. S5 , Supplementary Data 6 ). In some cases, these correlations confirm expectations and serve as a useful control for data quality. For example, iso-alpha acids, the bittering compounds in hops, strongly correlate with bitterness (Spearman’s rho=0.68), while ethanol and glycerol correlate with tasters’ perceptions of alcohol and body, the mouthfeel sensation of fullness (Spearman’s rho=0.82/0.62 and 0.72/0.57 respectively) and darker color from roasted malts is a good indication of malt perception (Spearman’s rho=0.54).

Heatmap colors indicate Spearman’s Rho. Axes are organized according to sensory categories (aroma, taste, mouthfeel, overall), chemical categories and chemical sources in beer (malt (blue), hops (green), yeast (red), wild flora (yellow), Others (black)). See Supplementary Data 6 for all correlation values.
Interestingly, for some relationships between chemical compounds and perceived flavor, correlations are weaker than expected. For example, the rose-smelling phenethyl acetate only weakly correlates with floral aroma. This hints at more complex relationships and interactions between compounds and suggests a need for a more complex model than simple correlations. Lastly, we uncovered unexpected correlations. For instance, the esters ethyl decanoate and ethyl octanoate appear to correlate slightly with hop perception and bitterness, possibly due to their fruity flavor. Iron is anti-correlated with hop aromas and bitterness, most likely because it is also anti-correlated with iso-alpha acids. This could be a sign of metal chelation of hop acids 61 , given that our analyses measure unbound hop acids and total iron content, or could result from the higher iron content in dark and Fruit beers, which typically have less hoppy and bitter flavors 62 .
Public consumer reviews complement expert panel data
To complement and expand the sensory data of our trained tasting panel, we collected 180,000 reviews of our 250 beers from the online consumer review platform RateBeer. This provided numerical scores for beer appearance, aroma, taste, palate, overall quality as well as the average overall score.
Public datasets are known to suffer from biases, such as price, cult status and psychological conformity towards previous ratings of a product. For example, prices correlate with appreciation scores for these online consumer reviews (rho=0.49, Supplementary Fig. S6 ), but not for our trained tasting panel (rho=0.19). This suggests that prices affect consumer appreciation, which has been reported in wine 63 , while blind tastings are unaffected. Moreover, we observe that some beer styles, like lagers and non-alcoholic beers, generally receive lower scores, reflecting that online reviewers are mostly beer aficionados with a preference for specialty beers over lager beers. In general, we find a modest correlation between our trained panel’s overall appreciation score and the online consumer appreciation scores (Fig. 3 , rho=0.29). Apart from the aforementioned biases in the online datasets, serving temperature, sample freshness and surroundings, which are all tightly controlled during the tasting panel sessions, can vary tremendously across online consumers and can further contribute to (among others, appreciation) differences between the two categories of tasters. Importantly, in contrast to the overall appreciation scores, for many sensory aspects the results from the professional panel correlated well with results obtained from RateBeer reviews. Correlations were highest for features that are relatively easy to recognize even for untrained tasters, like bitterness, sweetness, alcohol and malt aroma (Fig. 3 and below).
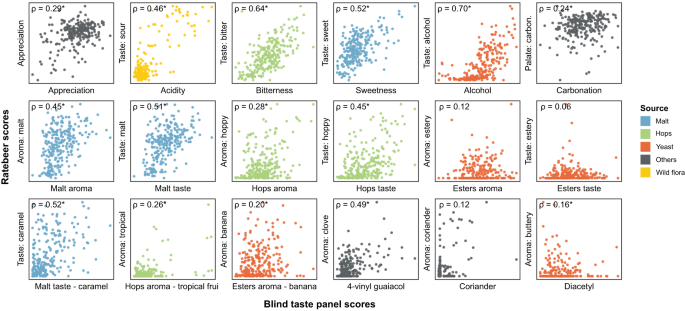
RateBeer text mining results can be found in Supplementary Data 7 . Rho values shown are Spearman correlation values, with asterisks indicating significant correlations ( p < 0.05, two-sided). All p values were smaller than 0.001, except for Esters aroma (0.0553), Esters taste (0.3275), Esters aroma—banana (0.0019), Coriander (0.0508) and Diacetyl (0.0134).
Besides collecting consumer appreciation from these online reviews, we developed automated text analysis tools to gather additional data from review texts (Supplementary Data 7 ). Processing review texts on the RateBeer database yielded comparable results to the scores given by the trained panel for many common sensory aspects, including acidity, bitterness, sweetness, alcohol, malt, and hop tastes (Fig. 3 ). This is in line with what would be expected, since these attributes require less training for accurate assessment and are less influenced by environmental factors such as temperature, serving glass and odors in the environment. Consumer reviews also correlate well with our trained panel for 4-vinyl guaiacol, a compound associated with a very characteristic aroma. By contrast, correlations for more specific aromas like ester, coriander or diacetyl are underrepresented in the online reviews, underscoring the importance of using a trained tasting panel and standardized tasting sheets with explicit factors to be scored for evaluating specific aspects of a beer. Taken together, our results suggest that public reviews are trustworthy for some, but not all, flavor features and can complement or substitute taste panel data for these sensory aspects.
Models can predict beer sensory profiles from chemical data
The rich datasets of chemical analyses, tasting panel assessments and public reviews gathered in the first part of this study provided us with a unique opportunity to develop predictive models that link chemical data to sensorial features. Given the complexity of beer flavor, basic statistical tools such as correlations or linear regression may not always be the most suitable for making accurate predictions. Instead, we applied different machine learning models that can model both simple linear and complex interactive relationships. Specifically, we constructed a set of regression models to predict (a) trained panel scores for beer flavor and quality and (b) public reviews’ appreciation scores from beer chemical profiles. We trained and tested 10 different models (Methods), 3 linear regression-based models (simple linear regression with first-order interactions (LR), lasso regression with first-order interactions (Lasso), partial least squares regressor (PLSR)), 5 decision tree models (AdaBoost regressor (ABR), extra trees (ET), gradient boosting regressor (GBR), random forest (RF) and XGBoost regressor (XGBR)), 1 support vector regression (SVR), and 1 artificial neural network (ANN) model.
To compare the performance of our machine learning models, the dataset was randomly split into a training and test set, stratified by beer style. After a model was trained on data in the training set, its performance was evaluated on its ability to predict the test dataset obtained from multi-output models (based on the coefficient of determination, see Methods). Additionally, individual-attribute models were ranked per descriptor and the average rank was calculated, as proposed by Korneva et al. 64 . Importantly, both ways of evaluating the models’ performance agreed in general. Performance of the different models varied (Table 1 ). It should be noted that all models perform better at predicting RateBeer results than results from our trained tasting panel. One reason could be that sensory data is inherently variable, and this variability is averaged out with the large number of public reviews from RateBeer. Additionally, all tree-based models perform better at predicting taste than aroma. Linear models (LR) performed particularly poorly, with negative R 2 values, due to severe overfitting (training set R 2 = 1). Overfitting is a common issue in linear models with many parameters and limited samples, especially with interaction terms further amplifying the number of parameters. L1 regularization (Lasso) successfully overcomes this overfitting, out-competing multiple tree-based models on the RateBeer dataset. Similarly, the dimensionality reduction of PLSR avoids overfitting and improves performance, to some extent. Still, tree-based models (ABR, ET, GBR, RF and XGBR) show the best performance, out-competing the linear models (LR, Lasso, PLSR) commonly used in sensory science 65 .
GBR models showed the best overall performance in predicting sensory responses from chemical information, with R 2 values up to 0.75 depending on the predicted sensory feature (Supplementary Table S4 ). The GBR models predict consumer appreciation (RateBeer) better than our trained panel’s appreciation (R 2 value of 0.67 compared to R 2 value of 0.09) (Supplementary Table S3 and Supplementary Table S4 ). ANN models showed intermediate performance, likely because neural networks typically perform best with larger datasets 66 . The SVR shows intermediate performance, mostly due to the weak predictions of specific attributes that lower the overall performance (Supplementary Table S4 ).
Model dissection identifies specific, unexpected compounds as drivers of consumer appreciation
Next, we leveraged our models to infer important contributors to sensory perception and consumer appreciation. Consumer preference is a crucial sensory aspects, because a product that shows low consumer appreciation scores often does not succeed commercially 25 . Additionally, the requirement for a large number of representative evaluators makes consumer trials one of the more costly and time-consuming aspects of product development. Hence, a model for predicting chemical drivers of overall appreciation would be a welcome addition to the available toolbox for food development and optimization.
Since GBR models on our RateBeer dataset showed the best overall performance, we focused on these models. Specifically, we used two approaches to identify important contributors. First, rankings of the most important predictors for each sensorial trait in the GBR models were obtained based on impurity-based feature importance (mean decrease in impurity). High-ranked parameters were hypothesized to be either the true causal chemical properties underlying the trait, to correlate with the actual causal properties, or to take part in sensory interactions affecting the trait 67 (Fig. 4A ). In a second approach, we used SHAP 68 to determine which parameters contributed most to the model for making predictions of consumer appreciation (Fig. 4B ). SHAP calculates parameter contributions to model predictions on a per-sample basis, which can be aggregated into an importance score.
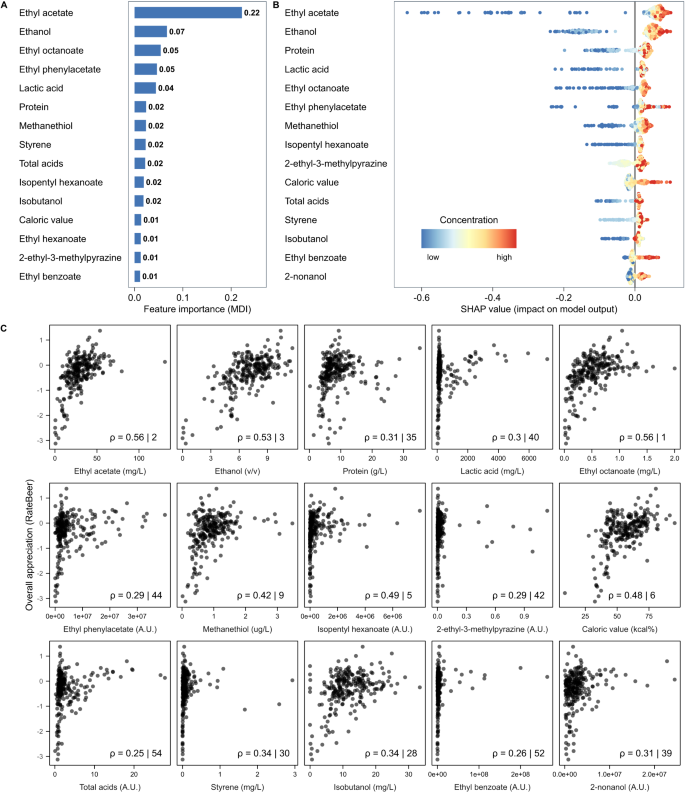
A The impurity-based feature importance (mean deviance in impurity, MDI) calculated from the Gradient Boosting Regression (GBR) model predicting RateBeer appreciation scores. The top 15 highest ranked chemical properties are shown. B SHAP summary plot for the top 15 parameters contributing to our GBR model. Each point on the graph represents a sample from our dataset. The color represents the concentration of that parameter, with bluer colors representing low values and redder colors representing higher values. Greater absolute values on the horizontal axis indicate a higher impact of the parameter on the prediction of the model. C Spearman correlations between the 15 most important chemical properties and consumer overall appreciation. Numbers indicate the Spearman Rho correlation coefficient, and the rank of this correlation compared to all other correlations. The top 15 important compounds were determined using SHAP (panel B).
Both approaches identified ethyl acetate as the most predictive parameter for beer appreciation (Fig. 4 ). Ethyl acetate is the most abundant ester in beer with a typical ‘fruity’, ‘solvent’ and ‘alcoholic’ flavor, but is often considered less important than other esters like isoamyl acetate. The second most important parameter identified by SHAP is ethanol, the most abundant beer compound after water. Apart from directly contributing to beer flavor and mouthfeel, ethanol drastically influences the physical properties of beer, dictating how easily volatile compounds escape the beer matrix to contribute to beer aroma 69 . Importantly, it should also be noted that the importance of ethanol for appreciation is likely inflated by the very low appreciation scores of non-alcoholic beers (Supplementary Fig. S4 ). Despite not often being considered a driver of beer appreciation, protein level also ranks highly in both approaches, possibly due to its effect on mouthfeel and body 70 . Lactic acid, which contributes to the tart taste of sour beers, is the fourth most important parameter identified by SHAP, possibly due to the generally high appreciation of sour beers in our dataset.
Interestingly, some of the most important predictive parameters for our model are not well-established as beer flavors or are even commonly regarded as being negative for beer quality. For example, our models identify methanethiol and ethyl phenyl acetate, an ester commonly linked to beer staling 71 , as a key factor contributing to beer appreciation. Although there is no doubt that high concentrations of these compounds are considered unpleasant, the positive effects of modest concentrations are not yet known 72 , 73 .
To compare our approach to conventional statistics, we evaluated how well the 15 most important SHAP-derived parameters correlate with consumer appreciation (Fig. 4C ). Interestingly, only 6 of the properties derived by SHAP rank amongst the top 15 most correlated parameters. For some chemical compounds, the correlations are so low that they would have likely been considered unimportant. For example, lactic acid, the fourth most important parameter, shows a bimodal distribution for appreciation, with sour beers forming a separate cluster, that is missed entirely by the Spearman correlation. Additionally, the correlation plots reveal outliers, emphasizing the need for robust analysis tools. Together, this highlights the need for alternative models, like the Gradient Boosting model, that better grasp the complexity of (beer) flavor.
Finally, to observe the relationships between these chemical properties and their predicted targets, partial dependence plots were constructed for the six most important predictors of consumer appreciation 74 , 75 , 76 (Supplementary Fig. S7 ). One-way partial dependence plots show how a change in concentration affects the predicted appreciation. These plots reveal an important limitation of our models: appreciation predictions remain constant at ever-increasing concentrations. This implies that once a threshold concentration is reached, further increasing the concentration does not affect appreciation. This is false, as it is well-documented that certain compounds become unpleasant at high concentrations, including ethyl acetate (‘nail polish’) 77 and methanethiol (‘sulfury’ and ‘rotten cabbage’) 78 . The inability of our models to grasp that flavor compounds have optimal levels, above which they become negative, is a consequence of working with commercial beer brands where (off-)flavors are rarely too high to negatively impact the product. The two-way partial dependence plots show how changing the concentration of two compounds influences predicted appreciation, visualizing their interactions (Supplementary Fig. S7 ). In our case, the top 5 parameters are dominated by additive or synergistic interactions, with high concentrations for both compounds resulting in the highest predicted appreciation.
To assess the robustness of our best-performing models and model predictions, we performed 100 iterations of the GBR, RF and ET models. In general, all iterations of the models yielded similar performance (Supplementary Fig. S8 ). Moreover, the main predictors (including the top predictors ethanol and ethyl acetate) remained virtually the same, especially for GBR and RF. For the iterations of the ET model, we did observe more variation in the top predictors, which is likely a consequence of the model’s inherent random architecture in combination with co-correlations between certain predictors. However, even in this case, several of the top predictors (ethanol and ethyl acetate) remain unchanged, although their rank in importance changes (Supplementary Fig. S8 ).
Next, we investigated if a combination of RateBeer and trained panel data into one consolidated dataset would lead to stronger models, under the hypothesis that such a model would suffer less from bias in the datasets. A GBR model was trained to predict appreciation on the combined dataset. This model underperformed compared to the RateBeer model, both in the native case and when including a dataset identifier (R 2 = 0.67, 0.26 and 0.42 respectively). For the latter, the dataset identifier is the most important feature (Supplementary Fig. S9 ), while most of the feature importance remains unchanged, with ethyl acetate and ethanol ranking highest, like in the original model trained only on RateBeer data. It seems that the large variation in the panel dataset introduces noise, weakening the models’ performances and reliability. In addition, it seems reasonable to assume that both datasets are fundamentally different, with the panel dataset obtained by blind tastings by a trained professional panel.
Lastly, we evaluated whether beer style identifiers would further enhance the model’s performance. A GBR model was trained with parameters that explicitly encoded the styles of the samples. This did not improve model performance (R2 = 0.66 with style information vs R2 = 0.67). The most important chemical features are consistent with the model trained without style information (eg. ethanol and ethyl acetate), and with the exception of the most preferred (strong ale) and least preferred (low/no-alcohol) styles, none of the styles were among the most important features (Supplementary Fig. S9 , Supplementary Table S5 and S6 ). This is likely due to a combination of style-specific chemical signatures, such as iso-alpha acids and lactic acid, that implicitly convey style information to the original models, as well as the low number of samples belonging to some styles, making it difficult for the model to learn style-specific patterns. Moreover, beer styles are not rigorously defined, with some styles overlapping in features and some beers being misattributed to a specific style, all of which leads to more noise in models that use style parameters.
Model validation
To test if our predictive models give insight into beer appreciation, we set up experiments aimed at improving existing commercial beers. We specifically selected overall appreciation as the trait to be examined because of its complexity and commercial relevance. Beer flavor comprises a complex bouquet rather than single aromas and tastes 53 . Hence, adding a single compound to the extent that a difference is noticeable may lead to an unbalanced, artificial flavor. Therefore, we evaluated the effect of combinations of compounds. Because Blond beers represent the most extensive style in our dataset, we selected a beer from this style as the starting material for these experiments (Beer 64 in Supplementary Data 1 ).
In the first set of experiments, we adjusted the concentrations of compounds that made up the most important predictors of overall appreciation (ethyl acetate, ethanol, lactic acid, ethyl phenyl acetate) together with correlated compounds (ethyl hexanoate, isoamyl acetate, glycerol), bringing them up to 95 th percentile ethanol-normalized concentrations (Methods) within the Blond group (‘Spiked’ concentration in Fig. 5A ). Compared to controls, the spiked beers were found to have significantly improved overall appreciation among trained panelists, with panelist noting increased intensity of ester flavors, sweetness, alcohol, and body fullness (Fig. 5B ). To disentangle the contribution of ethanol to these results, a second experiment was performed without the addition of ethanol. This resulted in a similar outcome, including increased perception of alcohol and overall appreciation.
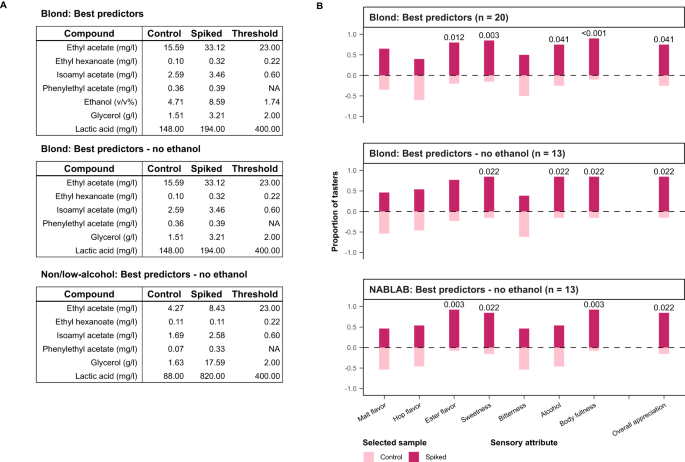
Adding the top chemical compounds, identified as best predictors of appreciation by our model, into poorly appreciated beers results in increased appreciation from our trained panel. Results of sensory tests between base beers and those spiked with compounds identified as the best predictors by the model. A Blond and Non/Low-alcohol (0.0% ABV) base beers were brought up to 95th-percentile ethanol-normalized concentrations within each style. B For each sensory attribute, tasters indicated the more intense sample and selected the sample they preferred. The numbers above the bars correspond to the p values that indicate significant changes in perceived flavor (two-sided binomial test: alpha 0.05, n = 20 or 13).
In a last experiment, we tested whether using the model’s predictions can boost the appreciation of a non-alcoholic beer (beer 223 in Supplementary Data 1 ). Again, the addition of a mixture of predicted compounds (omitting ethanol, in this case) resulted in a significant increase in appreciation, body, ester flavor and sweetness.
Predicting flavor and consumer appreciation from chemical composition is one of the ultimate goals of sensory science. A reliable, systematic and unbiased way to link chemical profiles to flavor and food appreciation would be a significant asset to the food and beverage industry. Such tools would substantially aid in quality control and recipe development, offer an efficient and cost-effective alternative to pilot studies and consumer trials and would ultimately allow food manufacturers to produce superior, tailor-made products that better meet the demands of specific consumer groups more efficiently.
A limited set of studies have previously tried, to varying degrees of success, to predict beer flavor and beer popularity based on (a limited set of) chemical compounds and flavors 79 , 80 . Current sensitive, high-throughput technologies allow measuring an unprecedented number of chemical compounds and properties in a large set of samples, yielding a dataset that can train models that help close the gaps between chemistry and flavor, even for a complex natural product like beer. To our knowledge, no previous research gathered data at this scale (250 samples, 226 chemical parameters, 50 sensory attributes and 5 consumer scores) to disentangle and validate the chemical aspects driving beer preference using various machine-learning techniques. We find that modern machine learning models outperform conventional statistical tools, such as correlations and linear models, and can successfully predict flavor appreciation from chemical composition. This could be attributed to the natural incorporation of interactions and non-linear or discontinuous effects in machine learning models, which are not easily grasped by the linear model architecture. While linear models and partial least squares regression represent the most widespread statistical approaches in sensory science, in part because they allow interpretation 65 , 81 , 82 , modern machine learning methods allow for building better predictive models while preserving the possibility to dissect and exploit the underlying patterns. Of the 10 different models we trained, tree-based models, such as our best performing GBR, showed the best overall performance in predicting sensory responses from chemical information, outcompeting artificial neural networks. This agrees with previous reports for models trained on tabular data 83 . Our results are in line with the findings of Colantonio et al. who also identified the gradient boosting architecture as performing best at predicting appreciation and flavor (of tomatoes and blueberries, in their specific study) 26 . Importantly, besides our larger experimental scale, we were able to directly confirm our models’ predictions in vivo.
Our study confirms that flavor compound concentration does not always correlate with perception, suggesting complex interactions that are often missed by more conventional statistics and simple models. Specifically, we find that tree-based algorithms may perform best in developing models that link complex food chemistry with aroma. Furthermore, we show that massive datasets of untrained consumer reviews provide a valuable source of data, that can complement or even replace trained tasting panels, especially for appreciation and basic flavors, such as sweetness and bitterness. This holds despite biases that are known to occur in such datasets, such as price or conformity bias. Moreover, GBR models predict taste better than aroma. This is likely because taste (e.g. bitterness) often directly relates to the corresponding chemical measurements (e.g., iso-alpha acids), whereas such a link is less clear for aromas, which often result from the interplay between multiple volatile compounds. We also find that our models are best at predicting acidity and alcohol, likely because there is a direct relation between the measured chemical compounds (acids and ethanol) and the corresponding perceived sensorial attribute (acidity and alcohol), and because even untrained consumers are generally able to recognize these flavors and aromas.
The predictions of our final models, trained on review data, hold even for blind tastings with small groups of trained tasters, as demonstrated by our ability to validate specific compounds as drivers of beer flavor and appreciation. Since adding a single compound to the extent of a noticeable difference may result in an unbalanced flavor profile, we specifically tested our identified key drivers as a combination of compounds. While this approach does not allow us to validate if a particular single compound would affect flavor and/or appreciation, our experiments do show that this combination of compounds increases consumer appreciation.
It is important to stress that, while it represents an important step forward, our approach still has several major limitations. A key weakness of the GBR model architecture is that amongst co-correlating variables, the largest main effect is consistently preferred for model building. As a result, co-correlating variables often have artificially low importance scores, both for impurity and SHAP-based methods, like we observed in the comparison to the more randomized Extra Trees models. This implies that chemicals identified as key drivers of a specific sensory feature by GBR might not be the true causative compounds, but rather co-correlate with the actual causative chemical. For example, the high importance of ethyl acetate could be (partially) attributed to the total ester content, ethanol or ethyl hexanoate (rho=0.77, rho=0.72 and rho=0.68), while ethyl phenylacetate could hide the importance of prenyl isobutyrate and ethyl benzoate (rho=0.77 and rho=0.76). Expanding our GBR model to include beer style as a parameter did not yield additional power or insight. This is likely due to style-specific chemical signatures, such as iso-alpha acids and lactic acid, that implicitly convey style information to the original model, as well as the smaller sample size per style, limiting the power to uncover style-specific patterns. This can be partly attributed to the curse of dimensionality, where the high number of parameters results in the models mainly incorporating single parameter effects, rather than complex interactions such as style-dependent effects 67 . A larger number of samples may overcome some of these limitations and offer more insight into style-specific effects. On the other hand, beer style is not a rigid scientific classification, and beers within one style often differ a lot, which further complicates the analysis of style as a model factor.
Our study is limited to beers from Belgian breweries. Although these beers cover a large portion of the beer styles available globally, some beer styles and consumer patterns may be missing, while other features might be overrepresented. For example, many Belgian ales exhibit yeast-driven flavor profiles, which is reflected in the chemical drivers of appreciation discovered by this study. In future work, expanding the scope to include diverse markets and beer styles could lead to the identification of even more drivers of appreciation and better models for special niche products that were not present in our beer set.
In addition to inherent limitations of GBR models, there are also some limitations associated with studying food aroma. Even if our chemical analyses measured most of the known aroma compounds, the total number of flavor compounds in complex foods like beer is still larger than the subset we were able to measure in this study. For example, hop-derived thiols, that influence flavor at very low concentrations, are notoriously difficult to measure in a high-throughput experiment. Moreover, consumer perception remains subjective and prone to biases that are difficult to avoid. It is also important to stress that the models are still immature and that more extensive datasets will be crucial for developing more complete models in the future. Besides more samples and parameters, our dataset does not include any demographic information about the tasters. Including such data could lead to better models that grasp external factors like age and culture. Another limitation is that our set of beers consists of high-quality end-products and lacks beers that are unfit for sale, which limits the current model in accurately predicting products that are appreciated very badly. Finally, while models could be readily applied in quality control, their use in sensory science and product development is restrained by their inability to discern causal relationships. Given that the models cannot distinguish compounds that genuinely drive consumer perception from those that merely correlate, validation experiments are essential to identify true causative compounds.
Despite the inherent limitations, dissection of our models enabled us to pinpoint specific molecules as potential drivers of beer aroma and consumer appreciation, including compounds that were unexpected and would not have been identified using standard approaches. Important drivers of beer appreciation uncovered by our models include protein levels, ethyl acetate, ethyl phenyl acetate and lactic acid. Currently, many brewers already use lactic acid to acidify their brewing water and ensure optimal pH for enzymatic activity during the mashing process. Our results suggest that adding lactic acid can also improve beer appreciation, although its individual effect remains to be tested. Interestingly, ethanol appears to be unnecessary to improve beer appreciation, both for blond beer and alcohol-free beer. Given the growing consumer interest in alcohol-free beer, with a predicted annual market growth of >7% 84 , it is relevant for brewers to know what compounds can further increase consumer appreciation of these beers. Hence, our model may readily provide avenues to further improve the flavor and consumer appreciation of both alcoholic and non-alcoholic beers, which is generally considered one of the key challenges for future beer production.
Whereas we see a direct implementation of our results for the development of superior alcohol-free beverages and other food products, our study can also serve as a stepping stone for the development of novel alcohol-containing beverages. We want to echo the growing body of scientific evidence for the negative effects of alcohol consumption, both on the individual level by the mutagenic, teratogenic and carcinogenic effects of ethanol 85 , 86 , as well as the burden on society caused by alcohol abuse and addiction. We encourage the use of our results for the production of healthier, tastier products, including novel and improved beverages with lower alcohol contents. Furthermore, we strongly discourage the use of these technologies to improve the appreciation or addictive properties of harmful substances.
The present work demonstrates that despite some important remaining hurdles, combining the latest developments in chemical analyses, sensory analysis and modern machine learning methods offers exciting avenues for food chemistry and engineering. Soon, these tools may provide solutions in quality control and recipe development, as well as new approaches to sensory science and flavor research.
Beer selection
250 commercial Belgian beers were selected to cover the broad diversity of beer styles and corresponding diversity in chemical composition and aroma. See Supplementary Fig. S1 .
Chemical dataset
Sample preparation.
Beers within their expiration date were purchased from commercial retailers. Samples were prepared in biological duplicates at room temperature, unless explicitly stated otherwise. Bottle pressure was measured with a manual pressure device (Steinfurth Mess-Systeme GmbH) and used to calculate CO 2 concentration. The beer was poured through two filter papers (Macherey-Nagel, 500713032 MN 713 ¼) to remove carbon dioxide and prevent spontaneous foaming. Samples were then prepared for measurements by targeted Headspace-Gas Chromatography-Flame Ionization Detector/Flame Photometric Detector (HS-GC-FID/FPD), Headspace-Solid Phase Microextraction-Gas Chromatography-Mass Spectrometry (HS-SPME-GC-MS), colorimetric analysis, enzymatic analysis, Near-Infrared (NIR) analysis, as described in the sections below. The mean values of biological duplicates are reported for each compound.
HS-GC-FID/FPD
HS-GC-FID/FPD (Shimadzu GC 2010 Plus) was used to measure higher alcohols, acetaldehyde, esters, 4-vinyl guaicol, and sulfur compounds. Each measurement comprised 5 ml of sample pipetted into a 20 ml glass vial containing 1.75 g NaCl (VWR, 27810.295). 100 µl of 2-heptanol (Sigma-Aldrich, H3003) (internal standard) solution in ethanol (Fisher Chemical, E/0650DF/C17) was added for a final concentration of 2.44 mg/L. Samples were flushed with nitrogen for 10 s, sealed with a silicone septum, stored at −80 °C and analyzed in batches of 20.
The GC was equipped with a DB-WAXetr column (length, 30 m; internal diameter, 0.32 mm; layer thickness, 0.50 µm; Agilent Technologies, Santa Clara, CA, USA) to the FID and an HP-5 column (length, 30 m; internal diameter, 0.25 mm; layer thickness, 0.25 µm; Agilent Technologies, Santa Clara, CA, USA) to the FPD. N 2 was used as the carrier gas. Samples were incubated for 20 min at 70 °C in the headspace autosampler (Flow rate, 35 cm/s; Injection volume, 1000 µL; Injection mode, split; Combi PAL autosampler, CTC analytics, Switzerland). The injector, FID and FPD temperatures were kept at 250 °C. The GC oven temperature was first held at 50 °C for 5 min and then allowed to rise to 80 °C at a rate of 5 °C/min, followed by a second ramp of 4 °C/min until 200 °C kept for 3 min and a final ramp of (4 °C/min) until 230 °C for 1 min. Results were analyzed with the GCSolution software version 2.4 (Shimadzu, Kyoto, Japan). The GC was calibrated with a 5% EtOH solution (VWR International) containing the volatiles under study (Supplementary Table S7 ).
HS-SPME-GC-MS
HS-SPME-GC-MS (Shimadzu GCMS-QP-2010 Ultra) was used to measure additional volatile compounds, mainly comprising terpenoids and esters. Samples were analyzed by HS-SPME using a triphase DVB/Carboxen/PDMS 50/30 μm SPME fiber (Supelco Co., Bellefonte, PA, USA) followed by gas chromatography (Thermo Fisher Scientific Trace 1300 series, USA) coupled to a mass spectrometer (Thermo Fisher Scientific ISQ series MS) equipped with a TriPlus RSH autosampler. 5 ml of degassed beer sample was placed in 20 ml vials containing 1.75 g NaCl (VWR, 27810.295). 5 µl internal standard mix was added, containing 2-heptanol (1 g/L) (Sigma-Aldrich, H3003), 4-fluorobenzaldehyde (1 g/L) (Sigma-Aldrich, 128376), 2,3-hexanedione (1 g/L) (Sigma-Aldrich, 144169) and guaiacol (1 g/L) (Sigma-Aldrich, W253200) in ethanol (Fisher Chemical, E/0650DF/C17). Each sample was incubated at 60 °C in the autosampler oven with constant agitation. After 5 min equilibration, the SPME fiber was exposed to the sample headspace for 30 min. The compounds trapped on the fiber were thermally desorbed in the injection port of the chromatograph by heating the fiber for 15 min at 270 °C.
The GC-MS was equipped with a low polarity RXi-5Sil MS column (length, 20 m; internal diameter, 0.18 mm; layer thickness, 0.18 µm; Restek, Bellefonte, PA, USA). Injection was performed in splitless mode at 320 °C, a split flow of 9 ml/min, a purge flow of 5 ml/min and an open valve time of 3 min. To obtain a pulsed injection, a programmed gas flow was used whereby the helium gas flow was set at 2.7 mL/min for 0.1 min, followed by a decrease in flow of 20 ml/min to the normal 0.9 mL/min. The temperature was first held at 30 °C for 3 min and then allowed to rise to 80 °C at a rate of 7 °C/min, followed by a second ramp of 2 °C/min till 125 °C and a final ramp of 8 °C/min with a final temperature of 270 °C.
Mass acquisition range was 33 to 550 amu at a scan rate of 5 scans/s. Electron impact ionization energy was 70 eV. The interface and ion source were kept at 275 °C and 250 °C, respectively. A mix of linear n-alkanes (from C7 to C40, Supelco Co.) was injected into the GC-MS under identical conditions to serve as external retention index markers. Identification and quantification of the compounds were performed using an in-house developed R script as described in Goelen et al. and Reher et al. 87 , 88 (for package information, see Supplementary Table S8 ). Briefly, chromatograms were analyzed using AMDIS (v2.71) 89 to separate overlapping peaks and obtain pure compound spectra. The NIST MS Search software (v2.0 g) in combination with the NIST2017, FFNSC3 and Adams4 libraries were used to manually identify the empirical spectra, taking into account the expected retention time. After background subtraction and correcting for retention time shifts between samples run on different days based on alkane ladders, compound elution profiles were extracted and integrated using a file with 284 target compounds of interest, which were either recovered in our identified AMDIS list of spectra or were known to occur in beer. Compound elution profiles were estimated for every peak in every chromatogram over a time-restricted window using weighted non-negative least square analysis after which peak areas were integrated 87 , 88 . Batch effect correction was performed by normalizing against the most stable internal standard compound, 4-fluorobenzaldehyde. Out of all 284 target compounds that were analyzed, 167 were visually judged to have reliable elution profiles and were used for final analysis.
Discrete photometric and enzymatic analysis
Discrete photometric and enzymatic analysis (Thermo Scientific TM Gallery TM Plus Beermaster Discrete Analyzer) was used to measure acetic acid, ammonia, beta-glucan, iso-alpha acids, color, sugars, glycerol, iron, pH, protein, and sulfite. 2 ml of sample volume was used for the analyses. Information regarding the reagents and standard solutions used for analyses and calibrations is included in Supplementary Table S7 and Supplementary Table S9 .
NIR analyses
NIR analysis (Anton Paar Alcolyzer Beer ME System) was used to measure ethanol. Measurements comprised 50 ml of sample, and a 10% EtOH solution was used for calibration.
Correlation calculations
Pairwise Spearman Rank correlations were calculated between all chemical properties.
Sensory dataset
Trained panel.
Our trained tasting panel consisted of volunteers who gave prior verbal informed consent. All compounds used for the validation experiment were of food-grade quality. The tasting sessions were approved by the Social and Societal Ethics Committee of the KU Leuven (G-2022-5677-R2(MAR)). All online reviewers agreed to the Terms and Conditions of the RateBeer website.
Sensory analysis was performed according to the American Society of Brewing Chemists (ASBC) Sensory Analysis Methods 90 . 30 volunteers were screened through a series of triangle tests. The sixteen most sensitive and consistent tasters were retained as taste panel members. The resulting panel was diverse in age [22–42, mean: 29], sex [56% male] and nationality [7 different countries]. The panel developed a consensus vocabulary to describe beer aroma, taste and mouthfeel. Panelists were trained to identify and score 50 different attributes, using a 7-point scale to rate attributes’ intensity. The scoring sheet is included as Supplementary Data 3 . Sensory assessments took place between 10–12 a.m. The beers were served in black-colored glasses. Per session, between 5 and 12 beers of the same style were tasted at 12 °C to 16 °C. Two reference beers were added to each set and indicated as ‘Reference 1 & 2’, allowing panel members to calibrate their ratings. Not all panelists were present at every tasting. Scores were scaled by standard deviation and mean-centered per taster. Values are represented as z-scores and clustered by Euclidean distance. Pairwise Spearman correlations were calculated between taste and aroma sensory attributes. Panel consistency was evaluated by repeating samples on different sessions and performing ANOVA to identify differences, using the ‘stats’ package (v4.2.2) in R (for package information, see Supplementary Table S8 ).
Online reviews from a public database
The ‘scrapy’ package in Python (v3.6) (for package information, see Supplementary Table S8 ). was used to collect 232,288 online reviews (mean=922, min=6, max=5343) from RateBeer, an online beer review database. Each review entry comprised 5 numerical scores (appearance, aroma, taste, palate and overall quality) and an optional review text. The total number of reviews per reviewer was collected separately. Numerical scores were scaled and centered per rater, and mean scores were calculated per beer.
For the review texts, the language was estimated using the packages ‘langdetect’ and ‘langid’ in Python. Reviews that were classified as English by both packages were kept. Reviewers with fewer than 100 entries overall were discarded. 181,025 reviews from >6000 reviewers from >40 countries remained. Text processing was done using the ‘nltk’ package in Python. Texts were corrected for slang and misspellings; proper nouns and rare words that are relevant to the beer context were specified and kept as-is (‘Chimay’,’Lambic’, etc.). A dictionary of semantically similar sensorial terms, for example ‘floral’ and ‘flower’, was created and collapsed together into one term. Words were stemmed and lemmatized to avoid identifying words such as ‘acid’ and ‘acidity’ as separate terms. Numbers and punctuation were removed.
Sentences from up to 50 randomly chosen reviews per beer were manually categorized according to the aspect of beer they describe (appearance, aroma, taste, palate, overall quality—not to be confused with the 5 numerical scores described above) or flagged as irrelevant if they contained no useful information. If a beer contained fewer than 50 reviews, all reviews were manually classified. This labeled data set was used to train a model that classified the rest of the sentences for all beers 91 . Sentences describing taste and aroma were extracted, and term frequency–inverse document frequency (TFIDF) was implemented to calculate enrichment scores for sensorial words per beer.
The sex of the tasting subject was not considered when building our sensory database. Instead, results from different panelists were averaged, both for our trained panel (56% male, 44% female) and the RateBeer reviews (70% male, 30% female for RateBeer as a whole).
Beer price collection and processing
Beer prices were collected from the following stores: Colruyt, Delhaize, Total Wine, BeerHawk, The Belgian Beer Shop, The Belgian Shop, and Beer of Belgium. Where applicable, prices were converted to Euros and normalized per liter. Spearman correlations were calculated between these prices and mean overall appreciation scores from RateBeer and the taste panel, respectively.
Pairwise Spearman Rank correlations were calculated between all sensory properties.
Machine learning models
Predictive modeling of sensory profiles from chemical data.
Regression models were constructed to predict (a) trained panel scores for beer flavors and quality from beer chemical profiles and (b) public reviews’ appreciation scores from beer chemical profiles. Z-scores were used to represent sensory attributes in both data sets. Chemical properties with log-normal distributions (Shapiro-Wilk test, p < 0.05 ) were log-transformed. Missing chemical measurements (0.1% of all data) were replaced with mean values per attribute. Observations from 250 beers were randomly separated into a training set (70%, 175 beers) and a test set (30%, 75 beers), stratified per beer style. Chemical measurements (p = 231) were normalized based on the training set average and standard deviation. In total, three linear regression-based models: linear regression with first-order interaction terms (LR), lasso regression with first-order interaction terms (Lasso) and partial least squares regression (PLSR); five decision tree models, Adaboost regressor (ABR), Extra Trees (ET), Gradient Boosting regressor (GBR), Random Forest (RF) and XGBoost regressor (XGBR); one support vector machine model (SVR) and one artificial neural network model (ANN) were trained. The models were implemented using the ‘scikit-learn’ package (v1.2.2) and ‘xgboost’ package (v1.7.3) in Python (v3.9.16). Models were trained, and hyperparameters optimized, using five-fold cross-validated grid search with the coefficient of determination (R 2 ) as the evaluation metric. The ANN (scikit-learn’s MLPRegressor) was optimized using Bayesian Tree-Structured Parzen Estimator optimization with the ‘Optuna’ Python package (v3.2.0). Individual models were trained per attribute, and a multi-output model was trained on all attributes simultaneously.
Model dissection
GBR was found to outperform other methods, resulting in models with the highest average R 2 values in both trained panel and public review data sets. Impurity-based rankings of the most important predictors for each predicted sensorial trait were obtained using the ‘scikit-learn’ package. To observe the relationships between these chemical properties and their predicted targets, partial dependence plots (PDP) were constructed for the six most important predictors of consumer appreciation 74 , 75 .
The ‘SHAP’ package in Python (v0.41.0) was implemented to provide an alternative ranking of predictor importance and to visualize the predictors’ effects as a function of their concentration 68 .
Validation of causal chemical properties
To validate the effects of the most important model features on predicted sensory attributes, beers were spiked with the chemical compounds identified by the models and descriptive sensory analyses were carried out according to the American Society of Brewing Chemists (ASBC) protocol 90 .
Compound spiking was done 30 min before tasting. Compounds were spiked into fresh beer bottles, that were immediately resealed and inverted three times. Fresh bottles of beer were opened for the same duration, resealed, and inverted thrice, to serve as controls. Pairs of spiked samples and controls were served simultaneously, chilled and in dark glasses as outlined in the Trained panel section above. Tasters were instructed to select the glass with the higher flavor intensity for each attribute (directional difference test 92 ) and to select the glass they prefer.
The final concentration after spiking was equal to the within-style average, after normalizing by ethanol concentration. This was done to ensure balanced flavor profiles in the final spiked beer. The same methods were applied to improve a non-alcoholic beer. Compounds were the following: ethyl acetate (Merck KGaA, W241415), ethyl hexanoate (Merck KGaA, W243906), isoamyl acetate (Merck KGaA, W205508), phenethyl acetate (Merck KGaA, W285706), ethanol (96%, Colruyt), glycerol (Merck KGaA, W252506), lactic acid (Merck KGaA, 261106).
Significant differences in preference or perceived intensity were determined by performing the two-sided binomial test on each attribute.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The data that support the findings of this work are available in the Supplementary Data files and have been deposited to Zenodo under accession code 10653704 93 . The RateBeer scores data are under restricted access, they are not publicly available as they are property of RateBeer (ZX Ventures, USA). Access can be obtained from the authors upon reasonable request and with permission of RateBeer (ZX Ventures, USA). Source data are provided with this paper.
Code availability
The code for training the machine learning models, analyzing the models, and generating the figures has been deposited to Zenodo under accession code 10653704 93 .
Tieman, D. et al. A chemical genetic roadmap to improved tomato flavor. Science 355 , 391–394 (2017).
Article ADS CAS PubMed Google Scholar
Plutowska, B. & Wardencki, W. Application of gas chromatography–olfactometry (GC–O) in analysis and quality assessment of alcoholic beverages – A review. Food Chem. 107 , 449–463 (2008).
Article CAS Google Scholar
Legin, A., Rudnitskaya, A., Seleznev, B. & Vlasov, Y. Electronic tongue for quality assessment of ethanol, vodka and eau-de-vie. Anal. Chim. Acta 534 , 129–135 (2005).
Loutfi, A., Coradeschi, S., Mani, G. K., Shankar, P. & Rayappan, J. B. B. Electronic noses for food quality: A review. J. Food Eng. 144 , 103–111 (2015).
Ahn, Y.-Y., Ahnert, S. E., Bagrow, J. P. & Barabási, A.-L. Flavor network and the principles of food pairing. Sci. Rep. 1 , 196 (2011).
Article CAS PubMed PubMed Central Google Scholar
Bartoshuk, L. M. & Klee, H. J. Better fruits and vegetables through sensory analysis. Curr. Biol. 23 , R374–R378 (2013).
Article CAS PubMed Google Scholar
Piggott, J. R. Design questions in sensory and consumer science. Food Qual. Prefer. 3293 , 217–220 (1995).
Article Google Scholar
Kermit, M. & Lengard, V. Assessing the performance of a sensory panel-panellist monitoring and tracking. J. Chemom. 19 , 154–161 (2005).
Cook, D. J., Hollowood, T. A., Linforth, R. S. T. & Taylor, A. J. Correlating instrumental measurements of texture and flavour release with human perception. Int. J. Food Sci. Technol. 40 , 631–641 (2005).
Chinchanachokchai, S., Thontirawong, P. & Chinchanachokchai, P. A tale of two recommender systems: The moderating role of consumer expertise on artificial intelligence based product recommendations. J. Retail. Consum. Serv. 61 , 1–12 (2021).
Ross, C. F. Sensory science at the human-machine interface. Trends Food Sci. Technol. 20 , 63–72 (2009).
Chambers, E. IV & Koppel, K. Associations of volatile compounds with sensory aroma and flavor: The complex nature of flavor. Molecules 18 , 4887–4905 (2013).
Pinu, F. R. Metabolomics—The new frontier in food safety and quality research. Food Res. Int. 72 , 80–81 (2015).
Danezis, G. P., Tsagkaris, A. S., Brusic, V. & Georgiou, C. A. Food authentication: state of the art and prospects. Curr. Opin. Food Sci. 10 , 22–31 (2016).
Shepherd, G. M. Smell images and the flavour system in the human brain. Nature 444 , 316–321 (2006).
Meilgaard, M. C. Prediction of flavor differences between beers from their chemical composition. J. Agric. Food Chem. 30 , 1009–1017 (1982).
Xu, L. et al. Widespread receptor-driven modulation in peripheral olfactory coding. Science 368 , eaaz5390 (2020).
Kupferschmidt, K. Following the flavor. Science 340 , 808–809 (2013).
Billesbølle, C. B. et al. Structural basis of odorant recognition by a human odorant receptor. Nature 615 , 742–749 (2023).
Article ADS PubMed PubMed Central Google Scholar
Smith, B. Perspective: Complexities of flavour. Nature 486 , S6–S6 (2012).
Pfister, P. et al. Odorant receptor inhibition is fundamental to odor encoding. Curr. Biol. 30 , 2574–2587 (2020).
Moskowitz, H. W., Kumaraiah, V., Sharma, K. N., Jacobs, H. L. & Sharma, S. D. Cross-cultural differences in simple taste preferences. Science 190 , 1217–1218 (1975).
Eriksson, N. et al. A genetic variant near olfactory receptor genes influences cilantro preference. Flavour 1 , 22 (2012).
Ferdenzi, C. et al. Variability of affective responses to odors: Culture, gender, and olfactory knowledge. Chem. Senses 38 , 175–186 (2013).
Article PubMed Google Scholar
Lawless, H. T. & Heymann, H. Sensory evaluation of food: Principles and practices. (Springer, New York, NY). https://doi.org/10.1007/978-1-4419-6488-5 (2010).
Colantonio, V. et al. Metabolomic selection for enhanced fruit flavor. Proc. Natl. Acad. Sci. 119 , e2115865119 (2022).
Fritz, F., Preissner, R. & Banerjee, P. VirtualTaste: a web server for the prediction of organoleptic properties of chemical compounds. Nucleic Acids Res 49 , W679–W684 (2021).
Tuwani, R., Wadhwa, S. & Bagler, G. BitterSweet: Building machine learning models for predicting the bitter and sweet taste of small molecules. Sci. Rep. 9 , 1–13 (2019).
Dagan-Wiener, A. et al. Bitter or not? BitterPredict, a tool for predicting taste from chemical structure. Sci. Rep. 7 , 1–13 (2017).
Pallante, L. et al. Toward a general and interpretable umami taste predictor using a multi-objective machine learning approach. Sci. Rep. 12 , 1–11 (2022).
Malavolta, M. et al. A survey on computational taste predictors. Eur. Food Res. Technol. 248 , 2215–2235 (2022).
Lee, B. K. et al. A principal odor map unifies diverse tasks in olfactory perception. Science 381 , 999–1006 (2023).
Mayhew, E. J. et al. Transport features predict if a molecule is odorous. Proc. Natl. Acad. Sci. 119 , e2116576119 (2022).
Niu, Y. et al. Sensory evaluation of the synergism among ester odorants in light aroma-type liquor by odor threshold, aroma intensity and flash GC electronic nose. Food Res. Int. 113 , 102–114 (2018).
Yu, P., Low, M. Y. & Zhou, W. Design of experiments and regression modelling in food flavour and sensory analysis: A review. Trends Food Sci. Technol. 71 , 202–215 (2018).
Oladokun, O. et al. The impact of hop bitter acid and polyphenol profiles on the perceived bitterness of beer. Food Chem. 205 , 212–220 (2016).
Linforth, R., Cabannes, M., Hewson, L., Yang, N. & Taylor, A. Effect of fat content on flavor delivery during consumption: An in vivo model. J. Agric. Food Chem. 58 , 6905–6911 (2010).
Guo, S., Na Jom, K. & Ge, Y. Influence of roasting condition on flavor profile of sunflower seeds: A flavoromics approach. Sci. Rep. 9 , 11295 (2019).
Ren, Q. et al. The changes of microbial community and flavor compound in the fermentation process of Chinese rice wine using Fagopyrum tataricum grain as feedstock. Sci. Rep. 9 , 3365 (2019).
Hastie, T., Friedman, J. & Tibshirani, R. The Elements of Statistical Learning. (Springer, New York, NY). https://doi.org/10.1007/978-0-387-21606-5 (2001).
Dietz, C., Cook, D., Huismann, M., Wilson, C. & Ford, R. The multisensory perception of hop essential oil: a review. J. Inst. Brew. 126 , 320–342 (2020).
CAS Google Scholar
Roncoroni, Miguel & Verstrepen, Kevin Joan. Belgian Beer: Tested and Tasted. (Lannoo, 2018).
Meilgaard, M. Flavor chemistry of beer: Part II: Flavor and threshold of 239 aroma volatiles. in (1975).
Bokulich, N. A. & Bamforth, C. W. The microbiology of malting and brewing. Microbiol. Mol. Biol. Rev. MMBR 77 , 157–172 (2013).
Dzialo, M. C., Park, R., Steensels, J., Lievens, B. & Verstrepen, K. J. Physiology, ecology and industrial applications of aroma formation in yeast. FEMS Microbiol. Rev. 41 , S95–S128 (2017).
Article PubMed PubMed Central Google Scholar
Datta, A. et al. Computer-aided food engineering. Nat. Food 3 , 894–904 (2022).
American Society of Brewing Chemists. Beer Methods. (American Society of Brewing Chemists, St. Paul, MN, U.S.A.).
Olaniran, A. O., Hiralal, L., Mokoena, M. P. & Pillay, B. Flavour-active volatile compounds in beer: production, regulation and control. J. Inst. Brew. 123 , 13–23 (2017).
Verstrepen, K. J. et al. Flavor-active esters: Adding fruitiness to beer. J. Biosci. Bioeng. 96 , 110–118 (2003).
Meilgaard, M. C. Flavour chemistry of beer. part I: flavour interaction between principal volatiles. Master Brew. Assoc. Am. Tech. Q 12 , 107–117 (1975).
Briggs, D. E., Boulton, C. A., Brookes, P. A. & Stevens, R. Brewing 227–254. (Woodhead Publishing). https://doi.org/10.1533/9781855739062.227 (2004).
Bossaert, S., Crauwels, S., De Rouck, G. & Lievens, B. The power of sour - A review: Old traditions, new opportunities. BrewingScience 72 , 78–88 (2019).
Google Scholar
Verstrepen, K. J. et al. Flavor active esters: Adding fruitiness to beer. J. Biosci. Bioeng. 96 , 110–118 (2003).
Snauwaert, I. et al. Microbial diversity and metabolite composition of Belgian red-brown acidic ales. Int. J. Food Microbiol. 221 , 1–11 (2016).
Spitaels, F. et al. The microbial diversity of traditional spontaneously fermented lambic beer. PLoS ONE 9 , e95384 (2014).
Blanco, C. A., Andrés-Iglesias, C. & Montero, O. Low-alcohol Beers: Flavor Compounds, Defects, and Improvement Strategies. Crit. Rev. Food Sci. Nutr. 56 , 1379–1388 (2016).
Jackowski, M. & Trusek, A. Non-Alcohol. beer Prod. – Overv. 20 , 32–38 (2018).
Takoi, K. et al. The contribution of geraniol metabolism to the citrus flavour of beer: Synergy of geraniol and β-citronellol under coexistence with excess linalool. J. Inst. Brew. 116 , 251–260 (2010).
Kroeze, J. H. & Bartoshuk, L. M. Bitterness suppression as revealed by split-tongue taste stimulation in humans. Physiol. Behav. 35 , 779–783 (1985).
Mennella, J. A. et al. A spoonful of sugar helps the medicine go down”: Bitter masking bysucrose among children and adults. Chem. Senses 40 , 17–25 (2015).
Wietstock, P., Kunz, T., Perreira, F. & Methner, F.-J. Metal chelation behavior of hop acids in buffered model systems. BrewingScience 69 , 56–63 (2016).
Sancho, D., Blanco, C. A., Caballero, I. & Pascual, A. Free iron in pale, dark and alcohol-free commercial lager beers. J. Sci. Food Agric. 91 , 1142–1147 (2011).
Rodrigues, H. & Parr, W. V. Contribution of cross-cultural studies to understanding wine appreciation: A review. Food Res. Int. 115 , 251–258 (2019).
Korneva, E. & Blockeel, H. Towards better evaluation of multi-target regression models. in ECML PKDD 2020 Workshops (eds. Koprinska, I. et al.) 353–362 (Springer International Publishing, Cham, 2020). https://doi.org/10.1007/978-3-030-65965-3_23 .
Gastón Ares. Mathematical and Statistical Methods in Food Science and Technology. (Wiley, 2013).
Grinsztajn, L., Oyallon, E. & Varoquaux, G. Why do tree-based models still outperform deep learning on tabular data? Preprint at http://arxiv.org/abs/2207.08815 (2022).
Gries, S. T. Statistics for Linguistics with R: A Practical Introduction. in Statistics for Linguistics with R (De Gruyter Mouton, 2021). https://doi.org/10.1515/9783110718256 .
Lundberg, S. M. et al. From local explanations to global understanding with explainable AI for trees. Nat. Mach. Intell. 2 , 56–67 (2020).
Ickes, C. M. & Cadwallader, K. R. Effects of ethanol on flavor perception in alcoholic beverages. Chemosens. Percept. 10 , 119–134 (2017).
Kato, M. et al. Influence of high molecular weight polypeptides on the mouthfeel of commercial beer. J. Inst. Brew. 127 , 27–40 (2021).
Wauters, R. et al. Novel Saccharomyces cerevisiae variants slow down the accumulation of staling aldehydes and improve beer shelf-life. Food Chem. 398 , 1–11 (2023).
Li, H., Jia, S. & Zhang, W. Rapid determination of low-level sulfur compounds in beer by headspace gas chromatography with a pulsed flame photometric detector. J. Am. Soc. Brew. Chem. 66 , 188–191 (2008).
Dercksen, A., Laurens, J., Torline, P., Axcell, B. C. & Rohwer, E. Quantitative analysis of volatile sulfur compounds in beer using a membrane extraction interface. J. Am. Soc. Brew. Chem. 54 , 228–233 (1996).
Molnar, C. Interpretable Machine Learning: A Guide for Making Black-Box Models Interpretable. (2020).
Zhao, Q. & Hastie, T. Causal interpretations of black-box models. J. Bus. Econ. Stat. Publ. Am. Stat. Assoc. 39 , 272–281 (2019).
Article MathSciNet Google Scholar
Hastie, T., Tibshirani, R. & Friedman, J. The Elements of Statistical Learning. (Springer, 2019).
Labrado, D. et al. Identification by NMR of key compounds present in beer distillates and residual phases after dealcoholization by vacuum distillation. J. Sci. Food Agric. 100 , 3971–3978 (2020).
Lusk, L. T., Kay, S. B., Porubcan, A. & Ryder, D. S. Key olfactory cues for beer oxidation. J. Am. Soc. Brew. Chem. 70 , 257–261 (2012).
Gonzalez Viejo, C., Torrico, D. D., Dunshea, F. R. & Fuentes, S. Development of artificial neural network models to assess beer acceptability based on sensory properties using a robotic pourer: A comparative model approach to achieve an artificial intelligence system. Beverages 5 , 33 (2019).
Gonzalez Viejo, C., Fuentes, S., Torrico, D. D., Godbole, A. & Dunshea, F. R. Chemical characterization of aromas in beer and their effect on consumers liking. Food Chem. 293 , 479–485 (2019).
Gilbert, J. L. et al. Identifying breeding priorities for blueberry flavor using biochemical, sensory, and genotype by environment analyses. PLOS ONE 10 , 1–21 (2015).
Goulet, C. et al. Role of an esterase in flavor volatile variation within the tomato clade. Proc. Natl. Acad. Sci. 109 , 19009–19014 (2012).
Article ADS CAS PubMed PubMed Central Google Scholar
Borisov, V. et al. Deep Neural Networks and Tabular Data: A Survey. IEEE Trans. Neural Netw. Learn. Syst. 1–21 https://doi.org/10.1109/TNNLS.2022.3229161 (2022).
Statista. Statista Consumer Market Outlook: Beer - Worldwide.
Seitz, H. K. & Stickel, F. Molecular mechanisms of alcoholmediated carcinogenesis. Nat. Rev. Cancer 7 , 599–612 (2007).
Voordeckers, K. et al. Ethanol exposure increases mutation rate through error-prone polymerases. Nat. Commun. 11 , 3664 (2020).
Goelen, T. et al. Bacterial phylogeny predicts volatile organic compound composition and olfactory response of an aphid parasitoid. Oikos 129 , 1415–1428 (2020).
Article ADS Google Scholar
Reher, T. et al. Evaluation of hop (Humulus lupulus) as a repellent for the management of Drosophila suzukii. Crop Prot. 124 , 104839 (2019).
Stein, S. E. An integrated method for spectrum extraction and compound identification from gas chromatography/mass spectrometry data. J. Am. Soc. Mass Spectrom. 10 , 770–781 (1999).
American Society of Brewing Chemists. Sensory Analysis Methods. (American Society of Brewing Chemists, St. Paul, MN, U.S.A., 1992).
McAuley, J., Leskovec, J. & Jurafsky, D. Learning Attitudes and Attributes from Multi-Aspect Reviews. Preprint at https://doi.org/10.48550/arXiv.1210.3926 (2012).
Meilgaard, M. C., Carr, B. T. & Carr, B. T. Sensory Evaluation Techniques. (CRC Press, Boca Raton). https://doi.org/10.1201/b16452 (2014).
Schreurs, M. et al. Data from: Predicting and improving complex beer flavor through machine learning. Zenodo https://doi.org/10.5281/zenodo.10653704 (2024).
Download references
Acknowledgements
We thank all lab members for their discussions and thank all tasting panel members for their contributions. Special thanks go out to Dr. Karin Voordeckers for her tremendous help in proofreading and improving the manuscript. M.S. was supported by a Baillet-Latour fellowship, L.C. acknowledges financial support from KU Leuven (C16/17/006), F.A.T. was supported by a PhD fellowship from FWO (1S08821N). Research in the lab of K.J.V. is supported by KU Leuven, FWO, VIB, VLAIO and the Brewing Science Serves Health Fund. Research in the lab of T.W. is supported by FWO (G.0A51.15) and KU Leuven (C16/17/006).
Author information
These authors contributed equally: Michiel Schreurs, Supinya Piampongsant, Miguel Roncoroni.
Authors and Affiliations
VIB—KU Leuven Center for Microbiology, Gaston Geenslaan 1, B-3001, Leuven, Belgium
Michiel Schreurs, Supinya Piampongsant, Miguel Roncoroni, Lloyd Cool, Beatriz Herrera-Malaver, Florian A. Theßeling & Kevin J. Verstrepen
CMPG Laboratory of Genetics and Genomics, KU Leuven, Gaston Geenslaan 1, B-3001, Leuven, Belgium
Leuven Institute for Beer Research (LIBR), Gaston Geenslaan 1, B-3001, Leuven, Belgium
Laboratory of Socioecology and Social Evolution, KU Leuven, Naamsestraat 59, B-3000, Leuven, Belgium
Lloyd Cool, Christophe Vanderaa & Tom Wenseleers
VIB Bioinformatics Core, VIB, Rijvisschestraat 120, B-9052, Ghent, Belgium
Łukasz Kreft & Alexander Botzki
AB InBev SA/NV, Brouwerijplein 1, B-3000, Leuven, Belgium
Philippe Malcorps & Luk Daenen
You can also search for this author in PubMed Google Scholar
Contributions
S.P., M.S. and K.J.V. conceived the experiments. S.P., M.S. and K.J.V. designed the experiments. S.P., M.S., M.R., B.H. and F.A.T. performed the experiments. S.P., M.S., L.C., C.V., L.K., A.B., P.M., L.D., T.W. and K.J.V. contributed analysis ideas. S.P., M.S., L.C., C.V., T.W. and K.J.V. analyzed the data. All authors contributed to writing the manuscript.
Corresponding author
Correspondence to Kevin J. Verstrepen .
Ethics declarations
Competing interests.
K.J.V. is affiliated with bar.on. The other authors declare no competing interests.
Peer review
Peer review information.
Nature Communications thanks Florian Bauer, Andrew John Macintosh and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information, peer review file, description of additional supplementary files, supplementary data 1, supplementary data 2, supplementary data 3, supplementary data 4, supplementary data 5, supplementary data 6, supplementary data 7, reporting summary, source data, source data, rights and permissions.
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/ .
Reprints and permissions
About this article
Cite this article.
Schreurs, M., Piampongsant, S., Roncoroni, M. et al. Predicting and improving complex beer flavor through machine learning. Nat Commun 15 , 2368 (2024). https://doi.org/10.1038/s41467-024-46346-0
Download citation
Received : 30 October 2023
Accepted : 21 February 2024
Published : 26 March 2024
DOI : https://doi.org/10.1038/s41467-024-46346-0
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
By submitting a comment you agree to abide by our Terms and Community Guidelines . If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.
Quick links
- Explore articles by subject
- Guide to authors
- Editorial policies
Sign up for the Nature Briefing: Translational Research newsletter — top stories in biotechnology, drug discovery and pharma.

IMAGES
VIDEO
COMMENTS
We include an overview of basic scientific writing principles, a detailed description of the sections of an original research article, and practical recommendations for selecting a journal and responding to peer review comments. ... Writing a scientific paper — a brief guide for new investigators. Vaccine 2017; 35 (5):722-8. 10.1016/j ...
Cooperative Institute for Research in Environmental Sciences, University of Colorado, UCB 334, Boulder, Colorado, 80309 USA. Search for more papers by this author. ... Regardless of the specific course being taught, this guide can be used as a reference when writing scientific papers, independent research projects, and laboratory reports. ...
We describe here the basic steps to follow in writing a scientific article. We outline the main sections that an average article should contain; the elements that should appear in these sections, and some pointers for making the overall result attractive and acceptable for publication. 1.
In each paragraph, the first sentence defines the context, the body contains the new idea and the final sentence offers a conclusion. For the whole paper, the introduction sets the context, the ...
The Scope of Scientific Writing 3 The Need for Clarity 3 Receiving the Signals 4 Understanding the Signals 4 Organization and Language in Scientific Writing 4 2 Historical Perspectives 6 The Early History 6 The Electronic Era 7 The IMRAD Story 8 3 Approaching a Writing Project 11 Establishing the Mind-Set 11 Preparing to Write 12 Doing the ...
Excellent science is an essential ingredient of any great research paper, but concise writing and a clear structure are also crucial. ... Scientific writing should always aim to be A, B and C ...
The present article, essentially based on TA Lang's guide for writing a scientific paper [ 1 ], will summarize the steps involved in the process of writing a scientific report and in increasing the likelihood of its acceptance. Figure 1. The Edwin Smith Papyrus (≈3000 BCE) Figure 2.
Delivered to your inbox every two weeks, the Writing Toolbox features practical advice and tools you can use to prepare a research manuscript for submission success and build your scientific writing skillset. Discover how to navigate the peer review and publishing process, beyond writing your article.
Then, writing the paper and getting it ready for submission may take me 3 to 6 months. I like separating the writing into three phases. The results and the methods go first, as this is where I write what was done and how, and what the outcomes were. In a second phase, I tackle the introduction and refine the results section with input from my ...
English Communication for Scientists, Unit 2.2. Effective writing is clear, accurate, and concise. When you are writing a paper, strive to write in a straightforward way. Construct sentences that ...
Module 1 • 3 hours to complete. In this section of the MOOC, you will learn what is necessary before writing a paper: the context in which the scientist is publishing. You will learn how to know your own community, through different exemples, and then we will present you how scientific journal and publication works.
This paper is a summary of the main and most recent recommendations in the scientific field on how to write a research paper to increase its impact. 1. Introduction. Papers are a crucial part of research; if research does not produce published papers, it remains incomplete [1].
The book covers all aspects of scientific writing from submission to publishing in detail. Written and edited by world leaders in the field. Chapters are easy to understand with essential contents for writing quality scientific research paper and easy to follow algorithms and key points in each chapter. Chapters highlight the importance of each ...
Writing is a big part of being a scientist, whether in the form of manuscripts, grants, reports, protocols, presentations, or even emails. However, many people look at writing as separate from science—a scientist writes, but scientists are not regarded as writers. 1 This outdated assertion means that writing and communication has been historically marginalized when it comes to training and ...
FORMAT FOR THE PAPER. Scientific research articles provide a method for scientists to communicate with other scientists about the results of their research. A standard format is used for these articles, in which the author presents the research in an orderly, logical manner. ... Victoria E. McMillan, Writing Papers in the Biological Sciences ...
Scientific writing is a technical form of writing that is designed to communicate scientific information to other scientists. Depending on the specific scientific genre—a journal article, a scientific poster, or a research proposal, for example—some aspects of the writing may change, such as its purpose, audience, or organization.Many aspects of scientific writing, however, vary little ...
This paper deals with the use of relative clauses in nineteenth-century scientific writing. For this purpose, a 30,000-word corpus was compiled, gathering texts from the fields of mechanics and ...
A scientific paper is a written report describing original research results. The format of a scientific paper has been defined by centuries of developing tradition, editorial practice, scientific ethics and the interplay with printing and publishing services. A scientific paper should have, in proper order, a Title, Abstract,
In conclusion, writing a scientific paper is a challenging task that requires a. combination of technical and creativ e skills, as well as knowledge of the specific. conven tions and guidelines ...
Scientific papers based on experimentation typically include five predominant sections: Abstract, Introduction, Methods, Results, and Discussion.This structure is a widely accepted approach to writing a research paper, and has specific sections that parallel the scientific method.
Scientific writing as a research skill. Scientific papers are often hard to read, even for specialists that work in the area. This matters because potential readers will often give up and do something else instead. And that means the paper will have less impact. The fact that many scientific papers are hard to read is surprising.
Scientific Papers. Scientific papers are for sharing your own original research work with other scientists or for reviewing the research conducted by others. As such, they are critical to the ...
It allows us to create a comprehensive and more complete dataset, essential for developing an accurate and robust model capable of extracting the desired information from the different sections of research papers. Finally, a dataset of over 4000 arXiv papers was extracted, focusing on the structure and impact of algorithmic research papers.
Research paper writing service is ready whenever you're ready. That's its main advantage. With 24/7 customer service there's no need to worry about time zones or late hours. That ensures a quick process and helps you to write a paper without any worries about deadlines. Happy clients will ensure you that this service is a life saver!
More from the blog. From Paper to Presentation: Redesigning Existing Figures for Slides January 15, 2024 Scientific figures do not equally suit all contexts. A figure designed for a paper will often be information-dense; multiple panels illustrate multiple ideas, multiple axes and color bars show the impact of numerous variables, annotations highlight specific caveats, and an extensive caption ...
To our knowledge, no previous research gathered data at this scale (250 samples, 226 chemical parameters, 50 sensory attributes and 5 consumer scores) to disentangle and validate the chemical ...