Thank you for visiting nature.com. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser (or turn off compatibility mode in Internet Explorer). In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript.
- View all journals

Genetics articles from across Nature Portfolio
Genetics is the branch of science concerned with genes, heredity, and variation in living organisms. It seeks to understand the process of trait inheritance from parents to offspring, including the molecular structure and function of genes, gene behaviour in the context of a cell or organism (e.g. dominance and epigenetics), gene distribution, and variation and change in populations.
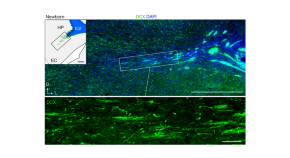
Neuron migration to brain regions key to memory and navigation continues into childhood
This study identifies a major migratory route for young neurons in the brains of young children. This route forms during pregnancy and links the birthplace of these nerve cells to their destination in highly interconnected brain regions that are responsible for memory and spatial processing.
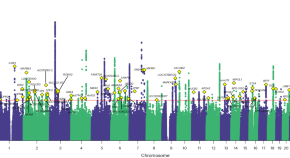
Deciphering the genetic architecture of pain intensity
Chronic pain is common, with more than one in five adult Americans reporting having pain daily or on most days. A multi-ancestry genomic analysis in 598,339 military veterans in the USA identifies 126 genetic variants associated with pain intensity, highlights shared genetic risk with substance use and psychiatric disorders, and reveals enrichment in GABAergic neurons as a key molecular contributor to experiencing pain.
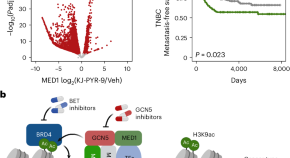
MYC activates transcriptional enhancers to drive cancer progression
We show that in addition to promoter activation, MYC drives cancer progression by activating transcriptional enhancers via a distinct mechanism. MYC cooperates with several other proteins at these cis -regulatory regions to change the epigenome and promote recruitment of RNA polymerase II and enhancer transcription.
Related Subjects
- Agricultural genetics
- Animal breeding
- Behavioural genetics
- Cancer genetics
- Cancer genomics
- Clinical genetics
- Consanguinity
- CRISPR-Cas systems
- Cytogenetics
- Development
- Epigenetics
- Epigenomics
- Evolutionary biology
- Functional genomics
- Gene expression
- Gene regulation
- Genetic association study
- Genetic hybridization
- Genetic interaction
- Genetic linkage study
- Genetic markers
- Genomic instability
- Heritable quantitative trait
- Immunogenetics
- Medical genetics
- Microbial genetics
- Neurodevelopmental disorders
- Plant breeding
- Plant genetics
- Population genetics
- Quantitative trait
- RNA splicing
Latest Research and Reviews
Genetic structure and origin of emu populations in Japanese farms inferred from large-scale SNP genotyping based on double-digest RAD-seq
- Yuichi Koshiishi
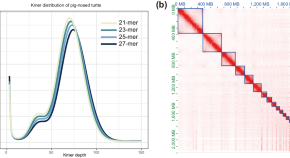
A chromosome-level genome assembly of the pig-nosed turtle ( Carettochelys insculpta )

A defined diet for pre-adult Drosophila melanogaster
- Felipe Martelli
- Annelise Quig
- Matthew D. W. Piper
Exogenous application of mycotoxin fusaric acid improve the morphological, cytogenetic, biochemical and anatomical parameters in salt (NaCl) stressed Allium cepa L.
- Kürşat Çavuşoğlu
- Dilek Çavuşoğlu
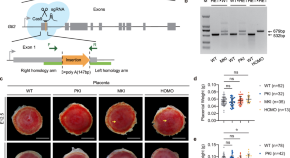
Maternal RNA transcription in Dlk1-Dio3 domain is critical for proper development of the mouse placental vasculature
Maternal RNA transcription plays an important role in maintaining vasculature in mouse placental development, associated with regulating gene expression, imprinting status and DNA methylation in the Dlk1-Dio3 imprinted domain.
- Ximeijia Zhang
- Hongjuan He
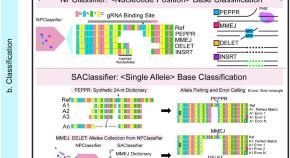
Developmental progression of DNA double-strand break repair deciphered by a single-allele resolution mutation classifier
DNA double-strand breaks (DSBs) are repaired by a hierarchically regulated network of pathways. Here, authors develop ICP for deciphering somatic DSB repair patterns in multicellular organisms and discover developmental regulation in flies and mosquitoes, enabling tracking of mutant alleles and interhomolog copying of gene cassettes.
News and Comment

Pregnancy advances your ‘biological’ age — but giving birth turns it back
Carrying a baby creates some of the same epigenetic patterns on DNA seen in older people.
- Saima Sidik

Global genomic diversity for All of Us
A publication in Nature reports the data release of around 245,000 clinical-grade whole-genome sequences as part of the NIH’s All of Us Research Programme. Several companion papers highlight the value of better capturing global genomic diversity.
Found: a skilled, courageous postdoctoral fellow
To select the best postdoctoral fellows for their labs, principal investigators assess scientific qualifications and more.
- Vivien Marx
The evolution of modifier genes
In this Journal Club, Yoav Ram recalls how he reconciled results from his own research with the reduction principle through the help of a paper published in PNAS by Altenberg et al.

Long-read sequencing provides insights into genetic influence
In this Tools of the Trade article, Trivett discusses the potential of long-read sequencing in generating high-quality reference genomes of animal models of cardiovascular disease.
- Cara Trivett
Quick links
- Explore articles by subject
- Guide to authors
- Editorial policies
- Share full article
Advertisement
Supported by
Scientists Finish the Human Genome at Last
The complete genome uncovered more than 100 new genes that are probably functional, and many new variants that may be linked to diseases.

By Carl Zimmer
Two decades after the draft sequence of the human genome was unveiled to great fanfare, a team of 99 scientists has finally deciphered the entire thing. They have filled in vast gaps and corrected a long list of errors in previous versions, giving us a new view of our DNA.
The consortium has posted six papers online in recent weeks in which they describe the full genome. These hard-sought data, now under review by scientific journals, will give scientists a deeper understanding of how DNA influences risks of disease, the scientists say, and how cells keep it in neatly organized chromosomes instead of molecular tangles.
For example, the researchers have uncovered more than 100 new genes that may be functional, and have identified millions of genetic variations between people. Some of those differences probably play a role in diseases.
For Nicolas Altemose, a postdoctoral researcher at the University of California, Berkeley, who worked on the team, the view of the complete human genome feels something like the close-up pictures of Pluto from the New Horizons space probe.
“You could see every crater, you could see every color, from something that we only had the blurriest understanding of before,” he said. “This has just been an absolute dream come true.”
Experts who were not involved in the project said it will enable scientists to explore the human genome in much greater detail. Large chunks of the genome that had been simply blank are now deciphered so clearly that scientists can start studying them in earnest.
“The fruit of this sequencing effort is amazing,” said Yukiko Yamashita, a developmental biologist at the Whitehead Institute for Biomedical Research at the Massachusetts Institute of Technology.
While scientists have known for decades that genes were spread across 23 pairs of chromosomes, these strange, wormlike microscopic structures remained largely a mystery.
By the late 1970s, scientists had gained the ability to pinpoint a few individual human genes and decode their sequence. But their tools were so crude that hunting down a single gene could take up an entire career.
Toward the end of the 20th Century, an international network of geneticists decided to try to sequence all the DNA in our chromosomes. The Human Genome Project was an audacious undertaking, given how much there was to sequence. Scientists knew that the twin strands of DNA in our cells contained roughly three billion pairs of letters — a text long enough to fill hundreds of books.
When that team began its work, the best technology the scientists could use sequenced bits of DNA just a few dozen letters, or bases, long. Researchers were left to put them together like the pieces of a vast jigsaw puzzle. To assemble the puzzle, they looked for fragments with identical ends, meaning that they came from overlapping portions of the genome. It took years for them to gradually assemble the sequenced fragments into larger swaths.
The White House announced in 2000 that scientists had finished the first draft of the human genome, and details of the project were published the following year. But long stretches of the genome remained unknown, while scientists struggled to figure out where millions of other bases belonged.
It turned out that the genome was a very hard puzzle to put together from small pieces. Many of our genes exist as multiple copies that are nearly identical to each other. Sometimes the different copies carry out different jobs. Other copies — known as pseudogenes — are disabled by mutations. A short fragment of DNA from one gene might fit just as well into the others.
And genes only make up a small percentage of the genome. The rest of it can be even more baffling . Much of the genome is made up of virus-like stretches of DNA that exist largely just to make new copies of themselves that get inserted back into the genome.
In the early 2000s, scientists got a little better at putting together the genome puzzle from its tiny pieces. They made more fragments, read them more accurately, and developed new computer programs to assemble them into bigger chunks of the genome.
Periodically, researchers would unveil the latest, best draft of the human genome — known as the reference genome. Scientists used the reference genome as a guide for their own sequencing efforts. For example, clinical geneticists would catalog disease-causing mutations by comparing genes from patients to the reference genome.
The newest reference genome came out in 2013. It was a lot better than the first draft, but it was a long way from complete. Eight percent of it was simply blank.
“There’s basically an entire human chromosome that had gone missing,” said Michael Schatz, a computational biologist at Johns Hopkins University.
In 2019, two scientists — Adam Phillippy, a computational biologist at the National Human Genome Research Institute, and Karen Miga, a geneticist at the University of California, Santa Cruz — founded the Telomere-to-Telomere Consortium to complete the genome.
Dr. Phillippy admitted that part of his motivation for such an audacious project was that the missing gaps annoyed him. “They were just really bugging me,” he said. “You take a beautiful landscape puzzle, pull out a hundred pieces, and look at it — that’s very bothersome to a perfectionist.”
Dr. Phillippy and Dr. Miga put out a call for scientists to join them to finish the puzzle. They ended up with 99 scientists working directly on sequencing the human genome, and dozens more pitching in to make sense of the data. The researchers worked remotely through the pandemic, coordinating their efforts over Slack, a messaging app.
“It was a surprisingly nice ant colony,” Dr. Miga said.
The consortium took advantage of new machines that can read stretches of DNA reaching tens of thousands of bases long. The researchers also invented techniques to figure out where particularly mysterious repeating sequences belonged in a genome.
All told, the scientists added or fixed more than 200 million base pairs in the reference genome. They can now say with confidence that the human genome measures 3.05 billion base pairs long.
Within those new sequences of DNA, the scientists discovered more than 2,000 new genes. Most appear to be disabled by mutations, but 115 of them look as if they can produce proteins — the function of which scientists may need years to figure out. The consortium now estimates that the human genome contains 19,969 protein-coding genes.
With a complete genome finally assembled, the researchers could take a better look at the variation in DNA from one person to the next. They discovered more than two million new spots in the genome where people differ. Using the new genome also helped them to avoid identifying disease-linked mutations where none actually exist.
“It’s a great advance for the field,” said Dr. Midhat Farooqui, the director of molecular oncology at Children’s Mercy, a hospital in Kansas City, Mo., who was not involved in the project.
Dr. Farooqi has started using the genome for his research into rare childhood diseases, aligning DNA from his patients against the newly filled gaps to search for mutations.
Switching to the new genome may be a challenge for many clinical labs, however. They’ll have to shift all of their information about the links between genes and diseases to a new map of the genome. “There will be a big effort, but it will take a couple years,” said Dr. Sharon Plon, a medical geneticist at Baylor College of Medicine in Houston.
Dr. Altemose plans on using the complete genome to explore a particularly mysterious region in each chromosome known as the centromere. Instead of storing genes, centromeres anchor proteins that move chromosomes around a cell as it divides. The centromere region contains thousands of repeated segments of DNA.
In their first look, Dr. Altemose and his colleagues were struck by how different centromere regions can be from one person to another. That observation suggests that centromeres have been evolving rapidly, as mutations insert new pieces of repeating DNA into the regions or cut other pieces out.
While some of this repeating DNA may play a role in pulling chromosomes apart, the researchers have also found new segments — some of them millions of bases long — that don’t appear to be involved. “We don’t know what they’re doing,” Dr. Altemose said.
But now that the empty zones of the genome are filled in, Dr. Altemose and his colleagues can study them up close. “I’m really excited moving forward to see all the things we can discover,” he said.
An earlier version of this article misstated when scientists first arrived at the correct number of human chromosomes. It was in the 1960s, not a century ago.
How we handle corrections
Carl Zimmer writes the “Matter” column. He is the author of fourteen books, including “Life's Edge: The Search For What It Means To Be Alive.” More about Carl Zimmer
The Mysteries and Wonders of Our DNA
Women are much more likely than men to have an array of so-called autoimmune diseases, like lupus and multiple sclerosis. A new study offers an explanation rooted in the X chromosome .
DNA fragments from thousands of years ago are providing insights into multiple sclerosis, diabetes, schizophrenia and other illnesses. Is this the future of medicine ?
A study of DNA from half a million volunteers found hundreds of mutations that could boost a young person’s fertility and that were linked to bodily damage later in life.
In the first effort of its kind, researchers now have linked DNA from 27 African Americans buried in the cemetery to nearly 42,000 living relatives .
Environmental DNA research has aided conservation, but scientists say its ability to glean information about humans poses dangers .
That person who looks just like you is not your twin. But if scientists compared your genomes, they might find a lot in common .

Essentials of Clinical Research pp 233–256 Cite as
Research Methods for Genetic Studies
- Sadeep Shrestha Ph.D., MHS, M.S. 2 &
- Donna K. Arnett Ph.D., M.S. 2
- First Online: 01 January 2014
4531 Accesses
This chapter introduces the basic concepts of fundamental methods for genotype-phenotype association studies and relevant issues in interpretation of genetic epidemiology studies to clinicians. An overview of genetic association studies is provided, which is the current state-of-the-art for clinical and translational genetics. Discussion of future directions in this field is also included.
- Genetic research
- Hardy Weinberg disequilibrium
- Familial aggregation
- Linkage disequilibrium
- Genome-wide association
This is a preview of subscription content, log in via an institution .
Buying options
- Available as PDF
- Read on any device
- Instant download
- Own it forever
- Available as EPUB and PDF
- Compact, lightweight edition
- Dispatched in 3 to 5 business days
- Free shipping worldwide - see info
- Durable hardcover edition
Tax calculation will be finalised at checkout
Purchases are for personal use only
Morton NE, Chung CS. Genetic epidemiology. New York: Academic; 1978.
Google Scholar
Mendel G. Versuche über Pflanzen-Hybriden. Verh. Naturforsch. Ver. Brünn 4: 3–47 (in English in 1901, J. R. Hortic. Soc. 26: 1–32)1866.
Kehrer-sawatzki H, Cooper DN. Copy number variation and disease. Basel/London: S Krager; 2009.
Bell J, Haldane JBS. The linkage between the genes for colour-blindness and haemophilia in man. Proc R Soc (Lond) B. 1937;123:119–50.
Article Google Scholar
Hartl DL, Clark AG. Principles of population genetics. Sunderland: Sinauer Associates; 2007.
Istrail S, Waterman M, Clark A. Computational methods for SNPs and haplotype inference. New York: Springer; 2004.
Book Google Scholar
Khoury MJ, Beaty TH, Cohen BH. Fundamental of genetic epidemiology. New York: Oxford University Press; 1993.
Ziegler A, Konig IR. Statistical approach to genetic epidemiology: concepts and applications. Weinheim: Wiley-VCH Verlag GmbH & Co. KGaA; 2006.
Knowler WC, Williams RC, Pettitt DJ, Steinberg AG. Gm3;5,13,14 and type 2 diabetes mellitus: an association in American Indians with genetic admixture. Am J Hum Genet. 1988;43(4):520–6.
CAS PubMed Central PubMed Google Scholar
Khoury MJ, Burke W, Thomson EJ. Genetics and public health in the 20th century. New York: Oxford University Press; 2000.
Additional References and Recommended Readings
1000 Genomes: http://www.1000genomes.org/
dbSNP National Center for Biotechnology Information (NCBI): http://www.ncbi.nlm.nih.gov/SNP/
ENCODE: https://genome.ucsc.edu/encode/
Human Genome Project: http://www.genome.gov/10001772
International HapMap Project: http://hapmap.ncbi.nlm.nih.gov/
Download references
Author information
Authors and affiliations.
Department of Epidemiology, School of Public Health, University of Alabama at Birmingham, Birmingham, AL, USA
Sadeep Shrestha Ph.D., MHS, M.S. & Donna K. Arnett Ph.D., M.S.
You can also search for this author in PubMed Google Scholar
Corresponding author
Correspondence to Sadeep Shrestha Ph.D., MHS, M.S. .
Editor information
Editors and affiliations.
Division of Preventive Medicine, University of Alabama at Birmingham, Birmingham, Alabama, USA
Stephen P. Glasser
Rights and permissions
Reprints and permissions
Copyright information
© 2014 Springer International Publishing Switzerland
About this chapter
Cite this chapter.
Shrestha, S., Arnett, D.K. (2014). Research Methods for Genetic Studies. In: Glasser, S. (eds) Essentials of Clinical Research. Springer, Cham. https://doi.org/10.1007/978-3-319-05470-4_11
Download citation
DOI : https://doi.org/10.1007/978-3-319-05470-4_11
Published : 30 April 2014
Publisher Name : Springer, Cham
Print ISBN : 978-3-319-05469-8
Online ISBN : 978-3-319-05470-4
eBook Packages : Medicine Medicine (R0)
Share this chapter
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Publish with us
Policies and ethics
- Find a journal
- Track your research
College of Biological Sciences
Discovery hints at genetic basis for the most challenging symptoms of schizophrenia.

Study suggests that current medications don’t target the right neural pathways to treat cognitive deficits often associated with the disease
- by Douglas Fox
- March 19, 2024
Our understanding of schizophrenia has increased greatly in recent years, as studies of large groups of people have identified a multitude of genetic variants that increase a person ’s risk of the disease. But each of those individual risk factors accounts for “ only a very minor amount of the overall risk , ” said Alex Nord, a professor of neurobiology, physiology and behavior in the College of Biological Sciences and the Center for Neuroscience.
Now a team of researchers co-led by Nord and graduate student Tracy Warren has connected genetic variants to the neural pathways that underlie some of the most challenging aspects of schizophrenia.
“ This is a new approach , ” said Nord . “ To have this level of detail is really powerful . ” The results were published March 15 in Molecular Psychiatry .
The work could one day lead to better diagnosis and treatment for the most devastating symptoms of schizophrenia – the ones most likely to leave a person homeless or unemployed – but least likely to respond to current medications.
A constellation of symptoms
Schizophrenia is often seen as a disorder of delusions and hallucinations, but the reality is more complex. People with the disease often suffer an array of symptoms, such as loss of motivation, inability to experience pleasure, social withdrawal, emotional flatness and even a tendency not to speak. Months or years can pass before a person is properly diagnosed and treatment is started — during which they can experience frayed relationships, lost jobs and deteriorating school performance. People with schizophrenia can also experience impaired cognitive control, making it difficult to organize their thoughts or direct their attention.
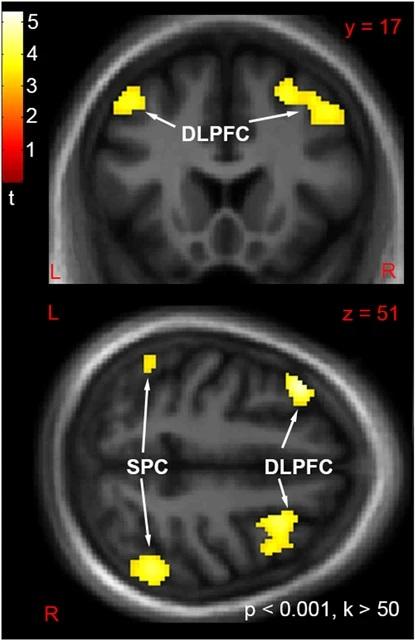
“ These symptoms are some of the strongest predictors of negative life outcomes ” such as unemployment, divorce, or homelessness, said Warren. Although existing medications can often curb hallucinations and delusions, “ currently, we don ’t have the drugs to fix these other symptoms . ”
Warren and Nord set out to study how genetic variants that are risk factors for schizophrenia actually contribute to the disease . They collaborated with Dr. Cameron Carter’s team at the Early Diagnosis and Preventive Treatment (EDAPT) Clinic, which offers psychosis care through the UC Davis School of Medicine and the Department of Psychiatry and Behavioral Sciences. Working with Carter and his team at EDAPT, Warren analyzed DNA from 205 patients and 115 healthy people. Carter’s team had examined these subjects in great detail, including interviews, cognitive tests, monitoring changes in their disease over time, and functional brain imaging to show how different brain areas worked together during mental tasks.
This rich patient and genetic dataset allowed Warren to attempt something new: examining genes linked to specific neural pathways to see if they are drivers of particular schizophrenia symptoms. Dr. Pak Sham at the University of Hong Kong was brought in as an expert on the genetic epidemiology of psychiatric disorders.
Working with Justin Tubbs, who was then a graduate student in Sham’s lab, Warren profiled these subjects against variants in more than 1,000 genes affecting four different neurotransmitter pathways — dopamine, GABA, glutamate, and serotonin.
Warren and Tubbs found that genetic variants in the glutamate and GABA pathways were strongly associated with deficits in cognitive control and social problems — a result that “ was really interesting , ” said Warren, because it hints at new strategies for treating the disease.
Treating the right neural pathway
Medications that are currently available for schizophrenia generally target dopamine and are usually effective in reducing hallucinations and delusions. But the new results suggest that cognitive problems — which greatly impact a person ’s life trajectory — are driven by neurotransmitter pathways that aren’t targeted by current medications.
“ This validates that we need to keep looking for treatments targeting these other pathways , ” Warren said.
When Warren divided the patients into clusters according to their individual groupings of disease symptoms and genetic variants, she found that patients with serious symptoms naturally separated into two subgroups — those who primarily experience cognitive problems, and those who primarily have hallucinations and delusions. Those with hallucinations and delusions usually respond well to existing medications, but those with cognitive problems do not.
Studies like this one could eventually pave the way toward more effective care for people with schizophrenia, Warren said. “ I would love to see people genotyped in the psychiatry clinic, have their neurotransmitter pathways examined, and their treatments selected based on this . ”
Nord believes that this approach, with early genetic testing, could also streamline the process of diagnosing schizophrenia and starting treatment. Diagnosis often consumes many months, allowing symptoms to worsen, yet the longer treatment is delayed, the more difficult it is to treat the disease.
“ Twenty years from now, we could potentially connect people with treatment earlier, before the symptoms become devastating , ” he said.

A leap of faith
For all this success, Nord admits that he was initially reluctant when Warren wanted to attempt this study — to use genetic data to connect neural pathways with symptoms. In a world where identifying the genetic risk variants required over 100,000 patients, it seemed far-fetched to think that they could take this next step with only 320 subjects.
“ I honestly didn ’t know if it would work , ” said Nord, who has spent two decades in research. “ Tracy really did drive this. It was a situation where the graduate student teaches us something . ”
Additional authors on the paper are: at UC Davis, Tyler A. Lesh, Mylena B. Corona, Sarvenaz S. Pakzad, Marina D. Albuquerque, and Praveena Singh; at UC Davis and Northwestern University, Vanessa Zarubin; at UC Davis and Washington University in St. Louis, Sarah J. Morse.
Media Resources
- Douglas Fox is a freelance science writer based in the Bay Area
- Association of neurotransmitter pathway polygenic risk with
- specific symptom profiles in psychosis ( Molecular Psychiatry 2024)
Primary Category
Secondary categories.
- Alzheimer's disease & dementia
- Arthritis & Rheumatism
- Attention deficit disorders
- Autism spectrum disorders
- Biomedical technology
- Diseases, Conditions, Syndromes
- Endocrinology & Metabolism
- Gastroenterology
- Gerontology & Geriatrics
- Health informatics
- Inflammatory disorders
- Medical economics
- Medical research
- Medications
- Neuroscience
- Obstetrics & gynaecology
- Oncology & Cancer
- Ophthalmology
- Overweight & Obesity
- Parkinson's & Movement disorders
- Psychology & Psychiatry
- Radiology & Imaging
- Sleep disorders
- Sports medicine & Kinesiology
- Vaccination
- Breast cancer
- Cardiovascular disease
- Chronic obstructive pulmonary disease
- Colon cancer
- Coronary artery disease
- Heart attack
- Heart disease
- High blood pressure
- Kidney disease
- Lung cancer
- Multiple sclerosis
- Myocardial infarction
- Ovarian cancer
- Post traumatic stress disorder
- Rheumatoid arthritis
- Schizophrenia
- Skin cancer
- Type 2 diabetes
- Full List »
share this!
March 19, 2024
This article has been reviewed according to Science X's editorial process and policies . Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
Genetic study finds epidemic of kuru likely led to migration of women in the Eastern Highlands of Papua New Guinea
by Cell Press

In the most comprehensive genetic study of the people living in the Eastern Highlands of Papua New Guinea to date, researchers from Papua New Guinea and the U.K. reveal the complex population structure and migration patterns of the communities in the region.
The results, published March 19 in The American Journal of Human Genetics , lay the foundation for future research on the fatal disease kuru—a disease transmitted during traditional anthropophagic mortuary practices—that devastated the area in the 20th century.
"We navigated one of the most complex landscapes possible in terms of geographical, cultural and linguistic diversity and set the scene for future genetic epidemiological work," said Liam Quinn, the paper's first author at the University College London and University of Copenhagen.
The Fore people, who until the mid-20th century were relatively isolated from the rest of the world in the Eastern Highlands of Papua New Guinea, had a tradition of mortuary feasts, during which they ate the bodies of their deceased loved ones as an expression of respect. However, a deadly disease was spreading to the people who participated in the feasts.
The illness particularly affected women and children who, as part of the mortuary rituals, consumed the infected tissue of the deceased individuals. The disease, known as kuru, led to a loss of motor coordination and balance, and then to a body tremor resulting in death. At the height of the epidemic, some villages had a significantly reduced female population.
"Out of respect for these communities, we do not use the word 'cannibalism' to describe this practice, but instead refer to it as mortuary feasts or anthropophagic mortuary practices," says Simon Mead, the paper's co-author at the UK Medical Research Council's Prion Unit at University College London.
"It was thought that anyone who attended a mortuary feast and ate infected brain tissue would die of kuru," says Mead. "But that was not correct. When the Papua New Guinea Institute of Medical Research (PNGIMR) field team interviewed elderly people in the affected population, it was clear that that there were people who participated in multiple mortuary feasts, but nevertheless survived."
The kuru epidemic dwindled over decades after anthropophagic mortuary practices were outlawed in the 1950s. However, researchers remained interested in the disease, particularly in understanding how some people lived after attending these feasts.
Kuru is a type of prion disease, which includes bovine spongiform encephalopathy—commonly known as mad cow disease—and its variant Creutzfeldt-Jakob disease, which sickened humans who consumed infected cattle, particularly in the UK. Prion diseases are incurable and fatal diseases caused by a change in the shape of the body's normal prion protein. Researchers are interested in exploring how humans can be protected from them.
Initial genetic research among the Fore people revealed that some of the elderly women who survived the feasts carried genetic variants in the gene that encodes prion proteins, which likely made them resistant to kuru.
"We found evidence that the Fore population was evolving to protect itself against the kuru epidemic, but this region had been ill-studied in the past, so we couldn't make confident inferences about evolution without a deeper knowledge of the genetics of the populations involved," Mead said.
Field staff from the affected and neighboring populations were recruited by the PNGIMR to collect genetic samples through long-term community participation, which were then analyzed with the assistance of Professor Simon Mead and colleagues in London and Copenhagen.
Genetic data from 943 individuals from 68 villages, representing 21 linguistic groups in the Eastern Highlands of Papua New Guinea, were analyzed, marking the most samples ever collected from the region. This data collection was supported by in-depth ethnographic studies of the traditional mortuary rites and oral history of the kuru-affected region.
"We're looking at a degree of sampling that provides resolution in the area that hasn't been achieved before in studies. This dataset has allowed us, for the first time, to ask some questions about the origins and interrelatedness of these communities, which traditionally had no written history, and how they've been shaped through longer periods of time," Quinn said.
The team used genome-wide genotyped data from all of the samples and compared the genomes of individuals from different villages and linguistic groups both within and beyond the Eastern Highlands. They found a strikingly high genetic distance between some linguistic groups in the region. The researchers suggested the difficult terrain, including mountains and fast-flowing rivers, may have contributed to these observations.
For example, the Anga and Fore linguistic groups were separated from one another by a fast-flowing river, but their genetic distance exceeds that observed between a population from the UK and one from Sri Lanka.
In addition, the researchers found a large influx of females migrating into the kuru-infected region over the years, challenging the notion that the kuru-infected groups became isolated from neighboring communities and excluded from the marital exchange because of their belief that the disease was caused by sorcery.
"Our findings provide evidence that supports another theory, which instead suggests that the flexible kinship systems of the Fore allowed them to intermarry with women from external communities despite fears of kuru. This likely helped the Fore to recover from the devastation caused by kuru, particularly to the female population of the Fore at the height of the epidemic," Quinn said.
Understanding these genetic distances and variations introduced by migration is crucial, the researchers said. Without these baseline data, scientists could falsely attribute genetic variants to kuru immunity when they were actually linked to these complex demographic reasons.
"This is another step in our long-term research endeavor to uncover how the Fore people and their neighbors survived kuru. It's also an important dataset that will help us further understand human history, because our knowledge about the populations in the region was scarce," Mead said.
Explore further
Feedback to editors
Researchers determine underlying mechanisms of inherited disorder that causes bone marrow failure
Mar 22, 2024
Intervention after first seizure may prevent long-term epilepsy

New genomic method offers diagnosis for patients with unexplained kidney failure
Researchers uncover protein interactions controlling fertility in female mice

Scientists close in on TB blood test that could detect millions of silent spreaders

New study reveals preventable-suicide risk profiles
Anti-inflammatory molecules show promise in reducing risks of further heart damage
A boost to biomedical research with statistical tools: From COVID-19 analysis to data management
UK study provides insights into COVID-19 vaccine uptake among children and young people
Researchers describe tools to better understand CaMKII, a protein involved in brain and heart disease
Related stories, study: no chronic wasting disease transmissibility in macaques.
Apr 25, 2018

Alzheimer's may have once spread from person to person, but the risk of that happening today is incredibly low
Feb 4, 2024

How prions invade the brain
Nov 29, 2018


Crucial step in formation of deadly brain diseases discovered
Mar 15, 2021

Huge genetic diversity among Papuan New Guinean peoples revealed
Sep 14, 2017

Oldest human remains from Puerto Rico reveal a complex cultural landscape since 1800 BC
Apr 26, 2023
Recommended for you
Accumulation of 'junk proteins' identified as one cause of aging and possible source of ALS

Researchers unlock secrets of birth defect origins, offering early detection and prevention strategies
Let us know if there is a problem with our content.
Use this form if you have come across a typo, inaccuracy or would like to send an edit request for the content on this page. For general inquiries, please use our contact form . For general feedback, use the public comments section below (please adhere to guidelines ).
Please select the most appropriate category to facilitate processing of your request
Thank you for taking time to provide your feedback to the editors.
Your feedback is important to us. However, we do not guarantee individual replies due to the high volume of messages.
E-mail the story
Your email address is used only to let the recipient know who sent the email. Neither your address nor the recipient's address will be used for any other purpose. The information you enter will appear in your e-mail message and is not retained by Medical Xpress in any form.
Newsletter sign up
Get weekly and/or daily updates delivered to your inbox. You can unsubscribe at any time and we'll never share your details to third parties.
More information Privacy policy
Donate and enjoy an ad-free experience
We keep our content available to everyone. Consider supporting Science X's mission by getting a premium account.
E-mail newsletter
- Introduction to Genomics
- Educational Resources
- Policy Issues in Genomics
- The Human Genome Project
- Funding Opportunities
- Funded Programs & Projects
- Division and Program Directors
- Scientific Program Analysts
- Contact by Research Area
- News & Events
- Research Areas
- Research investigators
- Research Projects
- Clinical Research
- Data Tools & Resources
- Genomics & Medicine
- Family Health History
- For Patients & Families
- For Health Professionals
- Jobs at NHGRI
- Training at NHGRI
- Funding for Research Training
- Professional Development Programs
- NHGRI Culture
- Social Media
- Broadcast Media
- Image Gallery
- Press Resources
- Organization
- NHGRI Director
- Mission & Vision
- Policies & Guidance
- Institute Advisors
- Strategic Vision
- Leadership Initiatives
- Diversity, Equity, and Inclusion
- Partner with NHGRI
- Staff Search
Participating in Genomics Research
Participating in genomics research is an opportunity to support scientific exploration of the genome and to help NIH in its mission to understand, prevent, detect and better treat disease. In some cases, research participants may also gain some individual benefit, such as diagnosis of a disease or access to a treatment in development.
Frequently Asked Questions
A wide range of genomic research studies need participants, each with different levels of involvement, and it is important to understand the scientific purpose of any study and any potential personal benefit or risk from participation. Consider the following list of questions before participating.
What is the purpose of the study?
The main reason to participate is to advance science, so make sure you know what the study is designed to investigate and that it's something you want to help support.
How much effort will I be expected to commit?
This very much varies from study to study. For any study you are considering, it is important that you understand what you will need to do to be a participant, and how much time it will take.
What healthcare will I receive as part of the study?
The primary purpose of research studies is to advance scientific knowledge, although you may receive some care as part of your participation. To receive clinical care, you should consult your regular physician.
How will participating affect my family?
This is one consideration that is especially important to consider for genomics research since genomic information is hereditary and may reveal information about relatives. Consider talking to your family members about your choice to participate in research. Also ask the researcher what information might be discovered in the course of the study and whether information might be returned to you or your family members. For example, genomic testing results could tell you information about disease risk, ancestry, and family relationships.
Is it safe to participate in genomics research?
Research participants are protected by a set of federal regulations, known as the Common Rule, which ensures that such research is conducted ethically. The Common Rule requires that researchers provide research participants with complete information about the study, and that they voluntarily consent to participation. The research team must inform you of any possible risks or benefits of participating in the study. You can also stop participating in the study at any time if you wish to do so. The Common Rule also ensures that research projects conducted by the NIH and studies funded by the NIH are reviewed by an Institutional Review Board comprised of scientists, bioethicists, and members of the public, to ensure the study is conducted ethically and that any risk to participants is minimized.
How will my privacy be protected?
Some genomics research is on 'de-identified' samples, meaning that information that identifies you, like your name and date of birth, is separated from your sample to protect your privacy. In other studies, scientists may need sensitive personal information to conduct their research; in that case, researchers will describe how they will protect that information during the consent process. Additionally, all NIH-funded studies that collect identifiable information are protected by Certficates of Confidentiality, or CoCs. A researcher whose study is covered by a CoC is prohibited from disclosing identifiable information about their research participants in legal proceedings, such as in the case of a subpoena.
There is also a Federal law called the Health Insurance Portability and Accountability Act (HIPAA) that restricts the sharing of medical information unless you have provided consent.
Might I be discriminated against as a result of information about my genome being revealed?
In addition to taking steps to protect the privacy of your genetic information, there is a Federal law called the Genetic Information Nondiscrimination Act (GINA) that prohibits employers and health insurers from discriminating you on the basis of your genetic information. Another Federal law called the Affordable Care Act (ACA) prevents health insurers from denying insurance to people with pre-existing conditions, including genetic conditions.
Get Involved
A key step to participating in research is identifying studies for which you are eligible. Below are resources providing detailed information on research involving research participants:
ClinicalTrials.gov
Around the Nation and Worldwide This is a searchable registry and results database of federally and privately supported clinical trials conducted in the United States and around the world. ClinicalTrials.gov gives you information about a trial's purpose, who may participate, locations, and phone numbers for more details. This information should be used in conjunction with advice from health care professionals. [ Tips for finding trials on ClinicalTrials.gov ]
NIH Clinical Research Studies At the NIH Clinical Center in Bethesda, Maryland The NIH maintains an online database of clinical research studies taking place at its Clinical Center, which is located on the NIH campus in Bethesda, Maryland. Studies are conducted by most of the institutes and centers across the NIH. The Clinical Center hosts a wide range of studies from rare diseases to chronic health conditions, as well as studies for healthy volunteers. Visitors can search by diagnosis, sign, symptom or other key words.
GenomeConnect
GenomeConnect is a unique patient registry that allows you to share your genetic and health information with researchers, clinicians, and others.
Click here for studies being conducted by NHGRI researchers.
ResearchMatch
This is an NIH-funded initiative to connect 1) people who are trying to find research studies, and 2) researchers seeking people to participate in their studies. It is a free, secure registry to make it easier for the public to volunteer and to become involved in clinical research studies that contribute to improved health in the future
UK Clinical Trials Gateway This database of clinical trials in the United Kingdom (UK) is curated by the UK National Health Service
International Clinical Trials Registry Platform
The World Health Organization (WHO) manages this database that lists clinical trials in countries all around the world
Additional Resources:
- Are Clinical Studies For You?
- FAQs about Clinical Studies
- Legal, Ethical, and Safety Issues
Last updated: January 10, 2018

Trans-ancestral Genome Wide Association Study of Sporadic and Recurrent Miscarriage
- Find this author on Google Scholar
- Find this author on PubMed
- Search for this author on this site
- ORCID record for Alexandra Reynoso
- For correspondence: [email protected]
- ORCID record for Priyanka Nandakumar
- ORCID record for Jingchunzi Shi
- ORCID record for Michael V. Holmes
- ORCID record for Stella Aslibekyan
- Info/History
- Supplementary material
- Preview PDF
Miscarriage is a common adverse pregnancy outcome, impacting approximately 15% of pregnancies. Herein, we present results of the largest trans-ancestral genome wide association study for miscarriage to date, based on 334,593 cases of sporadic, and 52,087 cases of recurrent miscarriage in the 23andMe, Inc. Research Cohort. We identified 10 novel genome-wide significant associations for sporadic miscarriage, and one for recurrent miscarriage. These loci mapped to genes with roles in neural development and telomere length, and to developmental disorders including autism spectrum disorder. Three variants, with similar directionality and magnitude of effect, replicated in a previously published GWAS. Using Mendelian randomization and triangulation, robust evidence was found for smoking causally increasing the risk of sporadic (genetic liability to ever vs never smoking: OR 1.13; 95%CI: 1.11-1.15; P=2.61e-42) and recurrent (OR 1.25; 95%CI: 1.21-1.30; P=5.47e-34) miscarriage, with moderate, yet triangulating, evidence identified for a potential etiological role of caffeine consumption.
Competing Interest Statement
Authors are employed by and hold stock with 23andMe, Inc.
Funding Statement
This study was funded by 23andMe, Inc.
Author Declarations
I confirm all relevant ethical guidelines have been followed, and any necessary IRB and/or ethics committee approvals have been obtained.
The details of the IRB/oversight body that provided approval or exemption for the research described are given below:
Individuals participating in this study provided informed consent and conducted survey participation online, under a protocol approved by the external AAHRPP-accredited IRB, Ethical & Independent Review Services (E&I Review, now Salus IRB).
I confirm that all necessary patient/participant consent has been obtained and the appropriate institutional forms have been archived, and that any patient/participant/sample identifiers included were not known to anyone (e.g., hospital staff, patients or participants themselves) outside the research group so cannot be used to identify individuals.
I understand that all clinical trials and any other prospective interventional studies must be registered with an ICMJE-approved registry, such as ClinicalTrials.gov. I confirm that any such study reported in the manuscript has been registered and the trial registration ID is provided (note: if posting a prospective study registered retrospectively, please provide a statement in the trial ID field explaining why the study was not registered in advance).
I have followed all appropriate research reporting guidelines, such as any relevant EQUATOR Network research reporting checklist(s) and other pertinent material, if applicable.
Author affiliations updated; Supplementary Material references updated.
Data Availability
The full GWAS summary statistics for the 23andMe discovery data set will be made available through 23andMe to qualified researchers under an agreement with 23andMe that protects the privacy of the 23andMe participants. Datasets will be made available at no cost for academic use. Please visit https://research.23andme.com/collaborate/#dataset-access/ for more information and to apply to access the data.
https://research.23andme.com/collaborate/#dataset-access/
View the discussion thread.
Supplementary Material
Thank you for your interest in spreading the word about medRxiv.
NOTE: Your email address is requested solely to identify you as the sender of this article.

Citation Manager Formats
- EndNote (tagged)
- EndNote 8 (xml)
- RefWorks Tagged
- Ref Manager
- Tweet Widget
- Facebook Like
- Google Plus One
Subject Area
- Genetic and Genomic Medicine
- Addiction Medicine (313)
- Allergy and Immunology (615)
- Anesthesia (157)
- Cardiovascular Medicine (2238)
- Dentistry and Oral Medicine (275)
- Dermatology (199)
- Emergency Medicine (367)
- Endocrinology (including Diabetes Mellitus and Metabolic Disease) (792)
- Epidemiology (11516)
- Forensic Medicine (10)
- Gastroenterology (674)
- Genetic and Genomic Medicine (3524)
- Geriatric Medicine (336)
- Health Economics (610)
- Health Informatics (2270)
- Health Policy (907)
- Health Systems and Quality Improvement (858)
- Hematology (332)
- HIV/AIDS (739)
- Infectious Diseases (except HIV/AIDS) (13103)
- Intensive Care and Critical Care Medicine (749)
- Medical Education (355)
- Medical Ethics (99)
- Nephrology (383)
- Neurology (3304)
- Nursing (189)
- Nutrition (502)
- Obstetrics and Gynecology (643)
- Occupational and Environmental Health (643)
- Oncology (1738)
- Ophthalmology (517)
- Orthopedics (206)
- Otolaryngology (283)
- Pain Medicine (220)
- Palliative Medicine (65)
- Pathology (433)
- Pediatrics (995)
- Pharmacology and Therapeutics (417)
- Primary Care Research (394)
- Psychiatry and Clinical Psychology (3029)
- Public and Global Health (5950)
- Radiology and Imaging (1211)
- Rehabilitation Medicine and Physical Therapy (710)
- Respiratory Medicine (803)
- Rheumatology (363)
- Sexual and Reproductive Health (343)
- Sports Medicine (307)
- Surgery (381)
- Toxicology (50)
- Transplantation (169)
- Urology (141)
Advertisement

Abstract 131: Utilizing human induced pluripotent stem cells (hiPSCs) as a model to study the genetic drivers of rhabdomyosarcoma
- Split-Screen
- Article contents
- Figures & tables
- Supplementary Data
- Peer Review
- Get Permissions
- Cite Icon Cite
- Search Site
- Version of Record March 22 2024
Celeste Romero , Yanbin Zheng , Lin Xu , Stephen Skapek; Abstract 131: Utilizing human induced pluripotent stem cells (hiPSCs) as a model to study the genetic drivers of rhabdomyosarcoma. Cancer Res 15 March 2024; 84 (6_Supplement): 131. https://doi.org/10.1158/1538-7445.AM2024-131
Download citation file:
- Ris (Zotero)
- Reference Manager
Rhabdomyosarcoma (RMS) is a childhood soft tissue sarcoma composed of skeletal myoblast-like cells that lack the capacity to differentiate despite their high expression of terminal differentiation markers like myogenin. There are two main types of RMS: fusion-positive RMS (FP-RMS), which is associated with a chromosomal translocation resulting in an oncogenic fusion protein (PAX3-FOXO1), and the more common form, fusion-negative RMS (FN-RMS) which lacks this translocation and is instead thought to be driven by numerous other oncogenes. Our lab used a Bayesian-based computational algorithm to analyze NGS sequencing data from 290 RMS samples and identified 25 potential genetic drivers of RMS. We aim to use induced pluripotent stem cells as a new model to study how these drivers, including the PAX3-FOXO1 fusion gene, influence RMS formation and progression. To this aim, we have successfully made a doxycycline inducible lentiviral vector containing the full-length sequence of human PAX3-FOXO1 which we used to ectopically express PAX3-FOXO1 in two well-characterized iPSC lines. We subcutaneously injected these iPSCs into NOD/SCID mice and treated them with doxycycline to form teratomas, non-malignant tumors containing tissue from all three germ layers. We hypothesized that PAX3-FOXO1 induction in these differentiating teratomas, may have transforming capabilities and may deregulate transcriptional programs to form FP-RMS-like tumors. We have found that PAX3-FOXO1 overexpressing teratomas grew faster and histologically look more “immature” than their control counterparts but do not seem to give rise to rhabdo-like tumors. We want to further understand what other critical factors are necessary to drive RMS formation by focusing on one of our drivers of interest EZH2, a histone methyltransferase implicated as either an oncogene or a tumor suppressor in certain forms of cancer. We have data suggesting that EZH2-dependent histone methylation activity may act as a proto-oncogene with PAX3-FOXO1 to silence the expression of key tumor suppressor genes CDKN2A and CDKN2B in RMS. To this aim, we reasoned that iPSCs differentiated into myogenic progenitors, one of the potential cells of origin of RMS, will be a novel way to study the role of oncogenes like PAX3-FOXO1 and EZH2 in RMS. We have derived myogenic progenitors from iPSCs (iPSC-MPCs) that express key myogenic factors like PAX7 and MYOD and differentiate and fuse to form mature myofibers. We plan to next explore how ectopic expression of EZH2 and/or PAX3-FOXO1 alters gene expression and histone methylation and whether these alterations can drive RMS formation in these iPSC-MPCs. Using this model system of iPSC-MPCs to drive expression of these oncogenes has the potential to offer new and exciting ways to identify the most important genes in driving cancer formation hopefully providing insight into better targets for treatment of RMS.
Citation Format: Celeste Romero, Yanbin Zheng, Lin Xu, Stephen Skapek. Utilizing human induced pluripotent stem cells (hiPSCs) as a model to study the genetic drivers of rhabdomyosarcoma [abstract]. In: Proceedings of the American Association for Cancer Research Annual Meeting 2024; Part 1 (Regular Abstracts); 2024 Apr 5-10; San Diego, CA. Philadelphia (PA): AACR; Cancer Res 2024;84(6_Suppl):Abstract nr 131.
Citing articles via
Email alerts.
- Online First
- Collections
- Online ISSN 1538-7445
- Print ISSN 0008-5472
AACR Journals
- Blood Cancer Discovery
- Cancer Discovery
- Cancer Epidemiology, Biomarkers & Prevention
- Cancer Immunology Research
- Cancer Prevention Research
- Cancer Research
- Cancer Research Communications
- Clinical Cancer Research
- Molecular Cancer Research
- Molecular Cancer Therapeutics
- Info for Advertisers
- Information for Institutions/Librarians
- Privacy Policy
- Copyright © 2023 by the American Association for Cancer Research.
This Feature Is Available To Subscribers Only
Sign In or Create an Account
An official website of the United States government
The .gov means it’s official. Federal government websites often end in .gov or .mil. Before sharing sensitive information, make sure you’re on a federal government site.
The site is secure. The https:// ensures that you are connecting to the official website and that any information you provide is encrypted and transmitted securely.
- Publications
- Account settings
Preview improvements coming to the PMC website in October 2024. Learn More or Try it out now .
- Advanced Search
- Journal List
- HHS Author Manuscripts

Recent advances in genetic studies of alcohol use disorders
Ishaan gupta.
1 Indian Institute of Technology, Delhi, India;
Rohan Dandavate
2 Indian Institute of Science Education and Research, Bhopal, India;
Pallavi Gupta
Viplav agrawal, manav kapoor.
3 Icahn School of Medicine at Mount Sinai, New york, USA
Purpose of review:
Alcohol use disorder (AUD) is a complex genetic disorder with very high heritability. This polygenic disorder not only results in increased morbidity and mortality, it is also a substantial social and economic burden on families and the nation. For past three decades, several genetic studies were conducted to identify genes and pathways associated with AUD. This review aims to summarize past efforts and recent advances in genetic association studies of AUD and related traits.
Recent findings:
Initial genetic association studies achieved a limted success and suffered from low power due to small sample sizes. AUD is a polygenic trait and data from several thousands individuals was required to identify the genetic factors of small effect sizes. The scenario changed recently with technological advances and significant reduction in cost of the genome wide association analyses (GWAS). This enabled researchers to generate genomic data on mega biobanks and cohorts with access to extensive clinical and non-clinical phenotypes. Public access to data from biobanks and collaborative efforts of researchers lead to identification of several novel loci associated with AUDs and related traits. Efforts are now underway to identify the causal variants under the GWAS loci to identify target genes and biological mechanisms underpining AUDs. Many GWAS variants occur in promoter or enhancer regions of the genes and are involved in regulation of gene expression of causal genes. This, large amounts of “omics” data from projects such as “ENCODE”, RoadMap and GTEx is also helping researchers to integrate “multi-omics” data to interpret functional significance of GWAS variants.
With current review, we aim to present the recent advances in genetic and molecular studies of AUDs. Recent successes in genetic studies of AUDs will definetely motivate researchers and lead to better therapeutic interventions for this complex disorder.
Introduction
Alcohol use disorder (AUD) is one of the most common and costly public health problems in the United States and throughout the world[ 1 , 2 ]. A person with AUD consumes alcohol in quantities that might be injurious to themselves and to people around them[ 2 , 3 ]. Infect, the physical and mental health issues associated with excessive consumption of alcohol are known for centuries. Worldwide, an estimated 20–30% cases of esophageal cancer, liver cancer, cirrhosis of the liver, homicide, epilepsy, and motor vehicle accidents can be attributed to excessive consumption of alcohol[ 4 ].
Factors influencing AUD
AUDs may transmit from one generation to other and have a high degree of familial association[ 5 ]. Many twin, adoption and family studies have provided consistent evidence for genetic predispositions to AUDs[ 6 – 8 ]. Heritable influences account for approximately 40% to 60% of the total variance to risk for alcoholism[ 6 , 7 ]. A host of other social, cultural and personal factors also influence the drinking behaviors of an individual[ 9 ]. It is also important to remember that availability of alcohol is the most important factor that influences the outcome of AUD[ 1 , 10 ]. A person who has never tried alcohol can not become alcohol dependent in spite of his/ her genetic susceptibility to alcoholism. Therefore it is pretty safe to assume that AUD results from a complex interplay of genetic susceptibility (genes associated with risk), environmental influences, and history of alcohol exposure[ 11 , 12 ]. These combined factors likely contribute to system-wide epigenetic alterations, post-translational modifications, and long-term allostatic changes in brain regions that underlie the alcohol use disorder[ 12 ].
Phenotypes/ traits to study AUD
Like many other complex traits, alcoholism appears to be clinically and etiologicaly hetrogenous[ 13 ]. This implies that there might be several steps and intermediate conditions in the development of AUD. Information about the underlying genetic factors that influence risk to AUD can be derived from multiple levels of AUD including amounts of drinks (Alcohol consumption), severity and symptoms of alcohol abuse and dependence. Commonly, genome wide association studies (GWAS) of alcoholism have focused on phenotypes based on the Diagnostic & Statistical Manual of Mental Disorders (DSM)[ 14 ]. In the 4th edition of the DSM (DSM-IV), alcohol dependence (AD) and abuse were considered as mutually exclusive diagnoses that together made up AUDs. DSM-V[ 14 , 15 ] on the other hand consolidated AD and abuse as a single disorder as AUD[ 15 ] , [ 16 ]. By considering AD and abuse under single umbrella increased the number of diagnosed subjects, but this number was still not large enough to design powerful GWAS studies. Therefore, many genetic studies of alcoholism also concentrated on nonclinical phenotypes, such as alcohol consumption and Alcohol Use Disorders Identification Test (AUDIT)[ 17 – 19 ], from large population based cohorts. The AUDIT, a 10-item, self-reported test was developed by the World Health Organization as a screen for hazardous and harmful drinking and can be used as a total (AUDIT-T), AUDIT-Consumption (AUDIT-C) and AUDIT-Problems (AUDIT-P) sub-scores.
Genetics of AUD
Given a large heritability for AUDs, sevral studies were conducted to identify the specific genes or genetic variations associated with AUD or related traits[ 8 , 20 – 22 ]. The gene identification efforts for AUDs can be divided into pre-GWAS and the GWAS era. The pre-GWAS era mainly focussed on genome-wide linkage and candidate gene studies[ 23 , 23 – 26 ]. Although, during this period many genes were nominated as the causal factors, a few genes actually showed consistent evidence of an association with AUD[ 23 , 27 ]. Even the initial GWAS studies suffered from low power due to smaller sample sizes and failed to identify credible evidence in favor of any particular gene[ 28 – 30 ]. Power of GWAS significantly improved in the past couple of years with the advent of large biobanks (UK Biobank) and the collaborative efforts of large consortiums (Psychiatric Genetics Consortium [PGC]). Following section will briefly describe the most successful and replicated findings in candidate gene studies before moving to recent advances in the field of AUD gene discovery.
Candidate gene studies of AUD and related traits
Most candidate genes selected for AUD genetic association studies can broadly be divided into two categories: 1) genes involved in central nervous system’s (CNS) response to alcohol or other addictive substances ( CHRNA5, GABRG1, GABRA2, OPRM1 etc.)[ 27 , 31 – 35 ] and 2) genes involved in alcohol metabolism ( ADH4, ADH1B, ALDH2 )[ 36 – 41 ]. Out of all candidate genes, role of ADH1B in AUD is very well established and replicated, particularly among populations of Asian descent[ 36 , 37 , 42 ]. However, variants in ADH1B are uncommon (~3–5%) in European Americans (EAs) and African Americans (AAs) [ 36 ]. This is the reason that a low frequency coding variant of ADH1B gene (rs1229984) was originally identified in Asian samples and was subsequently replicated in EAs and AAs in a large meta-analysis[ 36 ]. This single nucleotide change leads to replacement of Arg48 with His48 and results in a “atypical ADH” enzyme that exhibits several times higher catalytic activity than the normal enzyme. Increased accumulation of acetaldehyde from due to higher catalytic activity of “atypical ADH” is responsible for the flushing and severe symptoms of alcohol related sensitivity[ 37 , 40 ]. The intense aversive reaction to even smaller amounts of alcohol deter individuals from consuming large amounts of alcohol and protects them from developing AUD[ 36 , 37 , 40 , 42 ]. Indeed, the His48 allele was also found to be associated with lower alcohol consumption as measured by the subjects’ lifetime maximum alcohol consumption in a 24-hour period (β = −0.28 (95% CI −0.35, − 0.20), p value = 3.24 × 10 −13 )[ 36 ].
GWAS of AUD and related traits
GWASs represent the most recent paradigm shift for the gene discovery[ 43 ]. These hypothesis free genome scans allow interrogation of million of SNPs across thousands of genes at relatively modest cost[ 43 ]. Many GWAS’s of AUD, AD and alcohol consumption have been completed majorly in the European ancestry[ 28 – 30 , 44 – 53 ]. First reported GWAS study for AD (Treutlein et al. 2009) identified 2 genome-wide significant SNPs (rs7590720 and rs134694) in combined male only sample of 1,460 AD subjects and 2,332 controls[ 44 ]. The closest gene to the association signal PECR (peroxisomal trans-2-enoyl-CoA reductase) is involved in the metabolism of fatty acids. Two subsequent AD GWASs did not identify any novel genome-wide significant loci (COGA, SAGE)[ 29 , 54 ]. Further Heath and colleagues performed GWAS of quantitative indices of excessive alcohol consumption in moderate size cohort of Australian families but failed to identify any genome-wide significant variant[ 28 ]. Subsequent family and case control genome-wide efforts met with similar fate of limited success and non replication across different studies[ 52 , 55 ]. At this point in time researchers already realized that AUD is highly polygenic and sample size of individual cohort is not enough to identify the SNPs with very small effect sizes[ 28 ]. Availability of raw genotype data through dbGAP also made it a bit easier to meta-analyze the similarity ascertained cohorts with genome-wide SNP data. Schumann and colleagues[ 56 ] meta-analyzed 26,316 population based subjects and a follow-up sample of 21,185 EA subjects and identified variants in the autism susceptibility candidate 2 ( AUTS2 ) gene significantly associated with alcohol consumption (gms/ day/ kg of body weight). Subsequently, Kapoor and colleagues[ 49 ] meta-analyzed two large complementary and well-characterized EA cohorts assessed using the Semi-Structured Assessment for the Genetics of Alcoholism (SSAGA) and identified rs1229984 SNP in ADH1B to be genome-wide significantly associated with phenotype measuring maximum number of alcoholic drinks in 24 hour. This was the first alcoholism related GWAS that reported genome-wide significance at this locus[ 49 ].
The initial genome-wide meta-analysis had a few consistent findings, but the variants identified in these GWASs explained a very small proportion of heritability for alcohol related traits [ 28 , 49 , 57 , 58 ]. A very large sample size was needed to account for the missing heritability and it was a great deal of challenge to identify the well characterized large cohorts with AUD phenotypes. To address this challenge, recent genome-wide efforts focussed on larger sample sizes assembled via consortia-led meta-analyses ( Figure 1 and table 1 ). Researchers got access to alcohol consumption data for large number of individuals through UKBiobank and it finally provided necessary boost in gene identification efforts for alcholism. Clarke and colleagues[ 59 ] were the first group to take advantage of this cohort and reported genome-wide significant associations at 14 loci including ADH1B gene ( Table 1 ). Soon most of these initial findings were replicated in a very large meta-analysis of alcohol consumption of over 30 datasets (UKBibank, 23andMe and other GWAS) across nearly 1.2 million participants of European ancestry ( Table 1 ; Figure 1 )[ 60 ]. In this study, Liu and colleagues discovered 566 genetic variants in 406 loci associated with multiple stages of alcohol and tobacco use (initiation, cessation, and heaviness), with 150 loci evidencing pleiotropic association[ 60 ]. These results provided a very good starting point to evaluate the effects of these loci in model organisms and more precise measures of AUD.

Majority of genomic data for large alcohol consumption and AUD meta-analysis was either from UKBiobank or from Million Veterans Project. Several other cohorts from dbGAP also contributed to large sample size of alcohol consumption GWAS by Liu et al, 2019. Genome-wide data on 14,904 DSM-IV diagnosed AD individuals and 37,944 controls from 28 case/control and family-based studies were meta-analyzed for PGC’s AD GWAS.
List of recently published genome-wide association studies related to AUDs
Despite these advances with the GWASs of alcohol consumption, genetic studies of AUD and AD drew multiple challenges and limited success[ 45 ]. In a community based cohort (e.g. UKBiobank), it is relatively easy to derive the measure of alcohol consumption using number of alcoholic drinks consumed by an individual. But these large population based cohorts lack individuals diagnosed with AUD or AD. Substance use disorder working group of PGC (PGCSUD) tried to circumvent this problem by performing genome-wide meta-analysis of well characterized cohorts for DSM-IV diagnosed AD[ 45 ]. This meta-analysis identified, genome-wide significant effects of different ADH1B variants in European (rs1229984; P =9.8 ×10 –13 ) and African ancestries (rs2066702; P =2.2 ×10 –9 ). This study also found that the genetic underpinnings of DSM-IV diagnosed AD only partially overlap with those for alcohol consumption, underscoring the genetic distinction between pathological and nonpathological drinking behaviors[ 45 ]. Recently, Sanchez and colleagues also reported similar genetic differences among AUD (measured as AUDIT-P) and alcohol consumption (measured as AUDIT-C)[ 61 ]. In this meta-analysis (UKBiobank and 23andMe), AUDIT-P score showed a strong genetic correlation with alcohol dependence (PGC-SUD GWAS), while AUDIT-C score showed stronger genetic correlation with alcohol consumption[ 61 ]. Although, these genetics differences were not as apparent in African Americans in a recent large GWAS of AUD and AUDIT-C on individuals from Million Veteran Program (MVP)[ 62 ]. This GWAS reported many overlapping variants for AUDIT-C and AUD, with moderate-to-high genetic correlation between these traits (0.522 in EAs and 0.930 in AAs). Despite the significant genetic overlap between the AUDIT-C and AUD diagnosis, downstream analyses of MVP data revealed biologically meaningful points of divergence. Kranzler and colleagues[ 62 ] found that the polygenic risk score (PRS) calculated from AUD GWAS was significantly associated with tobacco use and multiple psychiatric disorders, whereas the AUDIT-C PRS did not show any association with these traits. These findings further confirmed that the AUD and alcohol consumption (measured by AUDIT-C in MVP) are genetically related but very distinct phenotypes.
Despite a significant boost in the number of genome-wide significant loci, variants identified in these large GWASs still explain a very small proportion of estimated genetic effect (heritability) for AUD and alcohol consumption. The SNP heritability estimates of AUDIT-C scores for all loci in MVP and the meta-analysis of the UKBiobank and 23andMe data ranged from 0.6 to 8 percent respectively[ 61 , 62 ]. Heritability estimates for AUD were slightly lower and ranged from 0.5 to 5.9 percent in MVP versus UKBiobank and 23andMe meta-analysis respectively[ 61 , 62 ]. These estimates are still significantly lower than the heritability estimates of AUD from twin and family studies. Given the heterogeneity in the diagnosis and polygenicity of this complex trait it seems that we are still short of required sample size to identify all the variants associated with disease. Some researchers working with other complex psychiatric traits argued that missing heritability can be explained by rare to low frequency variants of relatively large effect sizes. These rare variants can be identified by next generation sequencing in large cohorts. Contrary to expectations, recent sequencing studies for neurological disorders in moderate sample size are not much successful and have not yielded the intended results[ 63 , 64 ]. These studies concluded that the low frequency disease associated variants generally have low-moderate effect sizes and very large sample size is needed to identify these variants. Sequencing costs are moving down, but still these costs are prohibitive to perform a large whole genome sequencing study of AUD and other complex psychiatric disorders.
Functional significance of GWAS variants
Recent progress in GWAS of AUD has identified several variants across many loci that are significantly associated with alcoholism and related traits ( Table 2 ). Some variants in these loci result in amino acid changes (e.g. rs1229984 in ADH1B ) and known to alter the function of the gene to affect outcome of AUD. Most other AUD and alcohol consumption associated variants occur in intergenic or intronic regions and are not directly associated with protein coding changes[ 65 ]. Linkage disequilibrium (LD) at many loci span across thousands of variants and further makes it difficult to identify a causal SNP or genes associated with the disorder[ 66 ]. Thus, many AUD GWAS just annotated the nearest gene to the lead SNP as the susceptibility loci ( Table 2 ). Furthermore, due to different LD structure across datasets, many times individual study identified different lead SNPs and/ or different nearest genes within the same loci. Recent studies on psychiatric and neurological disorders showed that the most of genome-wide significant variants occur on active enhancers or promoters and might alter the expression level of nearby (cis) or distant (trans) gene[ 66 – 68 ]. These gene expression altering SNPs (expression quantitative loci or eQTLs) can be specific to a particular cell or tissue type. Several post-genomic helper tools such as FUMA[ 69 ] are available to functionally annotate the GWAS variants and to predict the functional consequence of a disease associated variant. More recent tools such as PrediXcan[ 70 ] and TWAS[ 71 ] can impute the genetic component of tissue-specific gene expression in GWAS datasets and help to connect changes in gene expression to trait outcome. Although size of eQTL and transcriptomic datasets can be a limiting factor to detect all functional association. Still initial application of PrediXcan has prioritized several genes (e.g. MAPT, CRHR1, FUT2, ADH1B, ADH4, ADH5, C1QTNF4, GCKR, DRD2 ) across different tissues [ 61 , 72 ]. In a recent study some of these genetic targets including ADH1B , GCKR , SLC39A8 and KLB have been shown to play a conserved role in phenotypic responses to alcohol in Caenorhabditis elegans [ 74 ]. Researchers are also using the post-mortem brain tissue from alcoholic subjects to understand the effect of long-term alcohol consumption on expression of genes. Recently, Kapoor and colleagues[ 73 ] have suggested that genes identified in alcohol related GWASs (AD and alcohol consumption) interact with genes affected by alcohol exposure and results in system wide changes across pathways and networks involved in alcoholism. Better understanding of these pathways will definitely result in better therapeutic interventions for AUD and problematic drinking[ 73 ].
Shared loci among GWAS for problematic drinking and alcohol consumption
Conclusions
For centuries, it was known that problematic drinking runs in families and genetic factors influence the etiology of AUD. Recent GWAS approaches have started elucidating the genetic loci related to AUD. At many GWAS loci strong LD extend to several hundred megabases and makes it difficult to identify the causal variant and candidate genes associated with alcoholism. Integration of genomic and transcriptomic data has opened the door for fine mapping and molecular genetic investigations into pathways and networks related to AUD. But there is still need of large scale “omics” data from diverse populations to effectively and accurately fine map the loci associated with alcoholism and other complex disorders. Many groups including Collaborative Studies of Genetics of Alcoholism (COGA) are generating “omics” data on African American and other diverse populations. Genomic data in diverse populations will also be useful for accurate disease risk prediction using polygenic risk scores. The future goal of precision medicine cannot be achieved without genetic association studies in diverse populations. Researchers from COGA are also generating transcriptomic and epigenomic data at single nuclei level from many different regions from post-mortem human brains of alcoholics and controls. Single nuclei transcriptomic analysis in human brain will be extremely useful to understand the role of various cellular lineages in development of AUDs. Genetic studies of AUD are advancing in the right direction and insights revealed will elucidate novel therapeutic targets resulting in better understanding of AUD biology.
Funding source:
This work is supported by National Institute on Alcohol Abuse and Alcoholism (R21AA026388 and U10AA008401).
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of a an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
Conflicts of interest
The authors declare that they have no conflicts of interest associated with this manuscript.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
References:
Crocodile study challenges assumptions of far-ranging behaviour finding Queensland crocs stay closer to home
"Redefining" research on Queensland crocodiles has found the majority remain closer to home than previously thought, with potential implications as to how the predators are managed in the Sunshine State.
In a joint study, the CSIRO and the Department of Environment and Science collected genetic samples from 1,176 saltwater crocodiles.
Scientists found 91 per cent were found less than 50 kilometres from where they hatched, with the vast majority in the far north of the state.
Scientists say the findings provide a new understanding of crocodile behaviour.
Study 'redefines' croc knowledge
Queensland's crocodile population of between 20,000 and 30,000 is split broadly between six populations, stretching from Rockhampton to the Gulf of Carpenteria, with 80 per cent north of Cooktown.
The Department of Environment and Science's Simon Booth said the study was significant.
"Although estuarine crocodiles are capable of moving anywhere along Queensland's coastline, the study has redefined what we know about Queensland's crocodile population," Dr Booth said.
"We had assumed Queensland crocodiles … were travelling long distances and moving between areas where they hatched and areas where they may establish territory.
"The evidence in this work suggests that about 90 per cent of crocodiles in Queensland are staying with 50 kilometres of where they hatched.
"This is contrary to the Northern Territory where they are seeing much higher populations, around 50 per cent, of crocodiles moving 200 kilometres or more."
Dr Booth said Queensland crocodiles travelled less than their NT counterparts because the lower population density meant they did not have to travel as far to establish their territory.
The NT also had more extensive flood plains, making it easier for crocodiles to travel.
Findings significant for management
Crocodile management is a contested issue in North Queensland, with regular concerns about the safety of people at popular water holes.
Safety concerns resurfaced recently when Townsville woman Alicia Mays unknowingly filmed a 3-metre crocodile while swimming with her children near Rollingstone Creek .
Three crocodiles were identified downstream from the creek.
The incident prompted Katter's Australian Party to call for the crocodiles to be removed, but authorities maintained the crocodiles' behaviour was not threatening.
The Queensland Department of Environment and Science said the findings would help the government continue to keep people safe while ensuring the ongoing survival of wild crocodiles.
Dr Booth said the research boosted understanding of how crocodiles behaved in each area they populated.
"It's very important as to understanding the impact that management actions can have, certainly for managing public safety around crocodiles, and also the conservation of the species," he said.
"In areas where we do have more crocodiles in the north, around Cairns, it's important to understand that our management actions, and that removing crocodiles from the wild, can have a heavy localised impact."
Dr Booth said it was important to remember the crocodile population density was much lower in Queensland than in the Northern Territory.
"Queensland is really not in the same realm as far as its crocodile population," he said.
- X (formerly Twitter)
Related Stories
A family came within metres of a crocodile in a swimming hole. so why didn't it attack.
Mother swimming with children unknowingly films 'sizeable' crocodile at popular Townsville swimming spot
DNA shows where crocodiles arriving into Darwin Harbour are migrating from
- Academic Research
- Airlie Beach
- Animal Science
- Conservation
- Crocodile Attacks
- Rockhampton

IMAGES
VIDEO
COMMENTS
RSS Feed. Genetics research is the scientific discipline concerned with the study of the role of genes in traits such as the development of disease. It has a key role in identifying potential ...
In 1987, the New York Times Magazine characterized the Human Genome Project as the "biggest, costliest, most provocative biomedical research project in history." 2 But in the years between the ...
Participants in clinical studies help current and future generations. Through these studies, researchers develop new diagnostic tests, more effective treatments, and better ways of managing diseases with genetic components. Participants in studies are actively involved in understanding their disorder and current research.
The future of genomics and genetics: What to expect. The study of genomics and genetics is rapidly advancing, meaning it will likely play an increasingly important role in human disease research and prevention. Some of the emerging trends in genomics and genetics include: Direct-to-consumer genetic testing
N Engl J Med 2024;390:1022-1028. Cerebral cavernous malformations occur in 0.5% of the population; 85% are sporadic, and 15% are familial or radiation-induced. Several genetic variants, including ...
RSS Feed. Genetics is the branch of science concerned with genes, heredity, and variation in living organisms. It seeks to understand the process of trait inheritance from parents to offspring ...
Each DNA strand is made of four chemical units, called nucleotide bases, which comprise the genetic "alphabet." The bases are adenine (A), thymine (T), guanine (G), and cytosine (C). Bases on opposite strands pair specifically: an A always pairs with a T; a C always pairs with a G. The order of the As, Ts, Cs and Gs determines the meaning of ...
At NHGRI, we are focused on advances in genomics research. Building on our leadership role in the initial sequencing of the human genome, we collaborate with the world's scientific and medical communities to enhance genomic technologies that accelerate breakthroughs and improve lives. By empowering and expanding the field of genomics, we can ...
In 2019, two scientists — Adam Phillippy, a computational biologist at the National Human Genome Research Institute, and Karen Miga, a geneticist at the University of California, Santa Cruz ...
A recent study in a neonatal intensive care unit in Texas studied outcomes for 278 infants who were referred for clinical exome sequencing, and found that 36.7% received a genetic diagnosis, and medical management was affected for 52% of infants with diagnoses [ 39 ].
Genetics Research is a fully open access journal providing a key forum for original research on all aspects of human and animal genetics, reporting key findings on genomes, genes, mutations, developmental, evolutionary, and population genetics as well as ethical, legal and social aspects. ... This study included 66 patients with CLL, diagnosed ...
Abstract. A century after the first twin and adoption studies of behavior in the 1920s, this review looks back on the journey and celebrates milestones in behavioral genetic research. After a whistle-stop tour of early quantitative genetic research and the parallel journey of molecular genetics, the travelogue focuses on the last fifty years.
Abstract. This chapter introduces the basic concepts of fundamental methods for genotype-phenotype association studies and relevant issues in interpretation of genetic epidemiology studies to clinicians. An overview of genetic association studies is provided, which is the current state-of-the-art for clinical and translational genetics.
Genetics Research. Genetics studies the genes and DNA coding that each person inherits from their parents and how these genes can lead to disease or better health. Some inherited genes cause diseases such as Tay-Sachs or sickle cell anemia, while mutations in genes can cause genetic disorders, such as Down syndrome.
The promise of genetic studies for improvements in human health has been widely espoused (for example, Hood, 1992; Fears and Poste, 1999; National Bioethics Advisory Commission, 1999), and genetic studies have achieved added significance as the end point of the Human Genome Project approaches (Pennisi, 1999). There is the strong sense that everyone at some point in the not too distant future ...
Our understanding of schizophrenia has increased greatly in recent years, as studies of large groups of people have identified a multitude of genetic variants that increase a person 's risk of the disease. But each of those individual risk factors accounts for " only a very minor amount of the overall risk, " said Alex Nord, a professor of neurobiology, physiology and behavior in the ...
Citation: Genetic study finds epidemic of kuru likely led to migration of women in the Eastern Highlands of Papua New Guinea (2024, March 19) retrieved 21 March 2024 from https://medicalxpress.com ...
Participating in genomics research is an opportunity to support scientific exploration of the genome and to help NIH in its mission to understand, prevent, detect and better treat disease. In some cases, research participants may also gain some individual benefit, such as diagnosis of a disease or access to a treatment in development.
Genetic counseling research, which strives to improve the quality and availability of genetic counseling services, benefits from these clinical data. Original articles contributed to a large proportion of the publication outputs and increased significantly over the last decade. ... Lastly, the research themes in this study were derived from ...
Miscarriage is a common adverse pregnancy outcome, impacting approximately 15% of pregnancies. Herein, we present results of the largest trans-ancestral genome wide association study for miscarriage to date, based on 334,593 cases of sporadic, and 52,087 cases of recurrent miscarriage in the 23andMe, Inc. Research Cohort. We identified 10 novel genome-wide significant associations for sporadic ...
Now a team of researchers co-led by Nord and graduate student Tracy Warren has connected genetic variants to the neural pathways that underlie some of the most challenging aspects of schizophrenia ...
Our lab used a Bayesian-based computational algorithm to analyze NGS sequencing data from 290 RMS samples and identified 25 potential genetic drivers of RMS. We aim to use induced pluripotent stem cells as a new model to study how these drivers, including the PAX3-FOXO1 fusion gene, influence RMS formation and progression.
Summary: With current review, we aim to present the recent advances in genetic and molecular studies of AUDs. Recent successes in genetic studies of AUDs will definetely motivate researchers and lead to better therapeutic interventions for this complex disorder. Keywords: alcohol use disorder, GWAS, SNPs, AUDIT scores, ADH1B, multi-omics.
In short: A study has found 91 per cent of Queensland crocodiles stay within 50km of where they hatched. The study challenged previous thoughts on crocodile behaviour in Queensland.