- Search by keyword
- Search by citation
Page 1 of 109

Based on molecular docking and real-time PCR technology, the two-component system Bae SR was investigated on the mechanism of drug resistance in CRAB
This study aimed to explore the role of the two-component system Bae SR in the mechanism of drug resistance in carbapenem-resistant A. baumannii (CRAB) using molecular docking and real-time polymerase chain react...
- View Full Text
Genomic characterization and related functional genes of γ- poly glutamic acid producing Bacillus subtilis
γ- poly glutamic acid (γ-PGA), a high molecular weight polymer, is synthesized by microorganisms and secreted into the extracellular space. Due to its excellent performance, γ-PGA has been widely used in vario...
In silico analysis of intestinal microbial instability and symptomatic markers in mice during the acute phase of severe burns
Severe burns may alter the stability of the intestinal flora and affect the patient’s recovery process. Understanding the characteristics of the gut microbiota in the acute phase of burns and their association...
The effect of white grub ( Maladera Verticalis ) larvae feeding on rhizosphere microbial characterization of aerobic rice ( Oryza sativa L.) in Puer City, Yunnan Province, China
Rhizosphere microorganisms are vital in plants’ growth and development and these beneficial microbes are recruited to the root-zone soil when experiencing various environmental stresses. However, the effect of...
Characterization of genes related to the efflux pump and porin in multidrug-resistant Escherichia coli strains isolated from patients with COVID-19 after secondary infection
Escherichia coli ( E. coli ) is a multidrug resistant opportunistic pathogen that can cause secondary bacterial infections in patients with COVID-19. This study aimed to determine the antimicrobial resistance profi...
Correction: Uncovering the complexity of childhood undernutrition through strain‑level analysis of the gut microbiome
The original article was published in BMC Microbiology 2024 24 :73
Optimization of fermentation conditions and medium components for chrysomycin a production by Streptomyces sp. 891-B6
Chrysomycin A (CA) is a promising antibiotic for treatment of Gram-positive bacterial infections and cancers. In order to enhance CA yield, optimization of fermentation conditions and medium components was car...
Integrative metagenomic analysis reveals distinct gut microbial signatures related to obesity
Obesity is a metabolic disorder closely associated with profound alterations in gut microbial composition. However, the dynamics of species composition and functional changes in the gut microbiome in obesity r...
Ultraviolet C inactivation of Coxiella burnetii for production of a structurally preserved whole cell vaccine antigen
Q fever, a worldwide-occurring zoonotic disease, can cause economic losses for public and veterinary health systems. Vaccines are not yet available worldwide and currently under development. In this regard, it...
Neutrophil extracellular traps formation: effect of Leishmania major promastigotes and salivary gland homogenates of Phlebotomus papatasi in human neutrophil culture
Leishmaniasis as a neglected tropical disease (NTD) is caused by the inoculation of Leishmania parasites via the bite of phlebotomine sand flies. After an infected bite, a series of innate and adaptive immune res...
Assessment of bacterial profile, antimicrobial susceptibility status, and associated factors of isolates among hospitalized patients at Dessie Comprehensive Specialized Hospital, Northeast Ethiopia
Antimicrobial resistant bacteria among hospitalized patients are becoming a major public health threat worldwide, mainly in developing countries. Infections by these multidrug resistant pathogens cause high ra...
A review of emerging health threats from zoonotic New World mammarenaviruses
Despite repeated spillover transmission and their potential to cause significant morbidity and mortality in human hosts, the New World mammarenaviruses remain largely understudied. These viruses are endemic to...
Impact of Limosilactobacillus fermentum probiotic treatment on gut microbiota composition in sahiwal calves with rotavirus diarrhea: A 16S metagenomic analysis study”
Diarrhea poses a major threat to bovine calves leading to mortality and economic losses. Among the causes of calf diarrhea, bovine rotavirus is a major etiological agent and may result in dysbiosis of gut micr...
Genetic characterizations of Cryptosporidium spp. from children with or without diarrhea in Wenzhou, China: high probability of zoonotic transmission
Cryptosporidium is a highly pathogenic parasite responsible for diarrhea in children worldwide. Here, the epidemiological status and genetic characteristics of Cryptosporidium in children with or without diarrhea...
Effect of stress urinary incontinence on vaginal microbial communities
Postpartum women often experience stress urinary incontinence (SUI) and vaginal microbial dysbiosis, which seriously affect women’s physical and mental health. Understanding the relationship between SUI and va...
Hospital distribution, seasonality, time trends and antifungal susceptibility profiles of all Aspergillus species isolated from clinical samples from 2015 to 2022 in a tertiary care hospital
Aspergillus species cause a variety of serious clinical conditions with increasing trend in antifungal resistance. The present study aimed at evaluating hospital epidemiology and antifungal susceptibility of all ...
Comparative analysis of proteomic adaptations in Enterococcus faecalis and Enterococcus faecium after long term bile acid exposure
All gastrointestinal pathogens, including Enterococcus faecalis and Enterococcus faecium , undergo adaptation processes during colonization and infection. In this study, we investigated by data-independent acquisi...
Influence of PhoPQ and PmrAB two component system alternations on colistin resistance from non- mcr colistin resistant clinical E. Coli strains
The current understanding of acquired chromosomal colistin resistance mechanisms in Enterobacterales primarily involves the disruption of the upstream PmrAB and PhoPQ two-component system (TCS) control caused by ...
Staphylococcus aureus foldase PrsA contributes to the folding and secretion of protein A
Staphylococcus aureus secretes a variety of proteins including virulence factors that cause diseases. PrsA, encoded by many Gram-positive bacteria, is a membrane-anchored lipoprotein that functions as a foldase t...
Transcriptional dynamics during Rhodococcus erythropolis infection with phage WC1
Belonging to the Actinobacteria phylum, members of the Rhodococcus genus thrive in soil, water, and even intracellularly. While most species are non-pathogenic, several cause respiratory disease in animals and, m...
A hypervirulent Acinetobacter baumannii strain has robust anti-phagocytosis ability
Acinetobacter baumannii ( A. baumannii ) is associated with both hospital-acquired infections (HAP) and community-acquired pneumonia (CAP). In this study, we present a novel CAP-associated A. baumannii (CAP-AB) str...
Restoration of gut dysbiosis through Clostridium butyricum and magnesium possibly balance blood glucose levels: an experimental study
Diabetes mellitus (DM) is a chronic metabolic disorder characterized by an elevated level of blood glucose due to the absence of insulin secretion, ineffectiveness, or lack of uptake of secreted insulin in the...
Bacillus subtilis SOM8 isolated from sesame oil meal for potential probiotic application in inhibiting human enteropathogens
While particular strains within the Bacillus species, such as Bacillus subtilis , have been commercially utilised as probiotics, it is critical to implement screening assays and evaluate the safety to identify pot...
Promiscuous, persistent and problematic: insights into current enterococcal genomics to guide therapeutic strategy
Vancomycin-resistant enterococci (VRE) are major opportunistic pathogens and the causative agents of serious diseases, such as urinary tract infections and endocarditis. VRE strains mainly include species of Ente...
Comparison of integron mediated antimicrobial resistance in clinical isolates of Escherichia coli from urinary and bacteremic sources
Antimicrobial resistance (AMR) is a global threat driven mainly by horizontal gene transfer (HGT) mechanisms through mobile genetic elements (MGEs) including integrons. The variable region (VR) of an integron ...
Structure predictions and functional insights into Amidase_3 domain containing N -acetylmuramyl-L-alanine amidases from Deinococcus indicus DR1
N -acetylmuramyl-L-alanine amidases are cell wall modifying enzymes that cleave the amide bond between the sugar residues and stem peptide in peptidoglycan. Amidases play a vital role in septal cell wall cleavag.....
Profile of non-tuberculous mycobacteria amongst tuberculosis presumptive people in Cameroon
Cameroon is a tuberculosis (TB) burden country with a 12% positivity among TB presumptive cases. Of the presumptive cases with a negative TB test, some are infected with Non-tuberculous Mycobacteria (NTM). How...
In vitro investigation of relationship between quorum-sensing system genes, biofilm forming ability, and drug resistance in clinical isolates of Pseudomonas aeruginosa
Pseudomonas aeruginosa is an opportunistic pathogen in the health-care systems and one of the primary causative agents with high mortality in hospitalized patients, particularly immunocompromised. The limitation ...
Relationship between heart failure and intestinal inflammation in infants with congenital heart disease
The association between heart failure (HF) and intestinal inflammation caused by a disturbed intestinal microbiota in infants with congenital heart disease (CHD) was investigated.
Clostridium butyricum inhibits the inflammation in children with primary nephrotic syndrome by regulating Th17/Tregs balance via gut-kidney axis
Primary nephrotic syndrome (PNS) is a common glomerular disease in children. Clostridium butyricum ( C. butyricum), a probiotic producing butyric acid, exerts effective in regulating inflammation. This study was d...
Human-derived bacterial strains mitigate colitis via modulating gut microbiota and repairing intestinal barrier function in mice
Unbalanced gut microbiota is considered as a pivotal etiological factor in colitis. Nevertheless, the precise influence of the endogenous gut microbiota composition on the therapeutic efficacy of probiotics in...
In vitro and in silico studies of enterobactin-inspired Ciprofloxacin and Fosfomycin first generation conjugates on the antibiotic resistant E. coli OQ866153
The emergence of antimicrobial resistance in bacterial pathogens is a growing concern worldwide due to its impact on the treatment of bacterial infections. The "Trojan Horse" strategy has been proposed as a po...
HPV-associated cervicovaginal microbiome and host metabolome characteristics
Cervicovaginal microbiome plays an important role in the persistence of HPV infection and subsequent disease development. However, cervicovaginal microbiota varied cross populations with different habits and r...
Transcriptomic and physiological analyses of Trichoderma citrinoviride HT-1 assisted phytoremediation of Cd contaminated water by Phragmites australis
Plant growth promoting microbe assisted phytoremediation is considered a more effective approach to rehabilitation than the single use of plants, but underlying mechanism is still unclear. In this study, we co...
Long-term push–pull cropping system shifts soil and maize-root microbiome diversity paving way to resilient farming system
The soil biota consists of a complex assembly of microbial communities and other organisms that vary significantly across farming systems, impacting soil health and plant productivity. Despite its importance, ...
Pretreatment with an antibiotics cocktail enhances the protective effect of probiotics by regulating SCFA metabolism and Th1/Th2/Th17 cell immune responses
Probiotics are a potentially effective therapy for inflammatory bowel disease (IBD); IBD is linked to impaired gut microbiota and intestinal immunity. However, the utilization of an antibiotic cocktail (Abx) p...
High-throughput sequencing reveals differences in microbial community structure and diversity in the conjunctival tissue of healthy and type 2 diabetic mice
To investigate the differences in bacterial and fungal community structure and diversity in conjunctival tissue of healthy and diabetic mice.
High prevalence of ST5-SCC mec II-t311 clone of methicillin-resistant Staphylococcus aureus isolated from bloodstream infections in East China
Methicillin-resistant Staphylococcus aureus (MRSA) is a challenging global health threat, resulting in significant morbidity and mortality worldwide. This study aims to determine the molecular characteristics and...
Characteristics of the oral and gastric microbiome in patients with early-stage intramucosal esophageal squamous cell carcinoma
Oral microbiome dysbacteriosis has been reported to be associated with the pathogenesis of advanced esophageal cancer. However, few studies investigated the potential role of oral and gastric microbiota in ear...
The potential role of Listeria monocytogenes in promoting colorectal adenocarcinoma tumorigenic process
Listeria monocytogenes is a foodborne pathogen, which can cause a severe illness, especially in people with a weakened immune system or comorbidities. The interactions between host and pathogens and between patho...
Evaluation of clinical characteristics and risk factors associated with Chlamydia psittaci infection based on metagenomic next-generation sequencing
Psittacosis is a zoonosis caused by Chlamydia psittaci , the clinical manifestations of Psittacosis range from mild illness to fulminant severe pneumonia with multiple organ failure. This study aimed to evaluate t...
Characterization of the broad-spectrum antibacterial activity of bacteriocin-like inhibitory substance-producing probiotics isolated from fermented foods
Antimicrobial peptides, such as bacteriocin, produced by probiotics have become a promising novel class of therapeutic agents for treating infectious diseases. Selected lactic acid bacteria (LAB) isolated from...
Metagenomic gut microbiome analysis of Japanese patients with multiple chemical sensitivity/idiopathic environmental intolerance
Although the pathology of multiple chemical sensitivity (MCS) is unknown, the central nervous system is reportedly involved. The gut microbiota is important in modifying central nervous system diseases. Howeve...
The effect of in vitro simulated colonic pH gradients on microbial activity and metabolite production using common prebiotics as substrates
The interplay between gut microbiota (GM) and the metabolization of dietary components leading to the production of short-chain fatty acids (SCFAs) is affected by a range of factors including colonic pH and ca...
Gut microbial network signatures of early colonizers in preterm neonates with extrauterine growth restriction
Extrauterine growth restriction (EUGR) represents a prevalent condition observed in preterm neonates, which poses potential adverse implications for both neonatal development and long-term health outcomes. The...
Early transcriptional changes of heavy metal resistance and multiple efflux genes in Xanthomonas campestris pv. campestris under copper and heavy metal ion stress
Copper-induced gene expression in Xanthomonas campestris pv. campestris (Xcc) is typically evaluated using targeted approaches involving qPCR. The global response to copper stress in Xcc and resistance to metal i...
Whole-genome sequencing and analysis of Chryseobacterium arthrosphaerae from Rana nigromaculata
Chryseobacterium arthrosphaerae strain FS91703 was isolated from Rana nigromaculata in our previous study. To investigate the genomic characteristics, pathogenicity-related genes, antimicrobial resistance, and ph...
Changes in the nasopharyngeal and oropharyngeal microbiota in pediatric obstructive sleep apnea before and after surgery: a prospective study
To explore the changes and potential mechanisms of microbiome in different parts of the upper airway in the development of pediatric OSA and observe the impact of surgical intervention on oral microbiome for p...
Unveiling biological activities of biosynthesized starch/silver-selenium nanocomposite using Cladosporium cladosporioides CBS 174.62
Microbial cells capability to tolerate the effect of various antimicrobial classes represent a major worldwide health concern. The flexible and multi-components nanocomposites have enhanced physicochemical cha...
Characterization of the major autolysin ( AtlC ) of Staphylococcus carnosus
Autolysis by cellular peptidoglycan hydrolases (PGH) is a well-known phenomenon in bacteria. During food fermentation, autolysis of starter cultures can exert an accelerating effect, as described in many studi...
Important information
Editorial board
For authors
For editorial board members
For reviewers
- Manuscript editing services
Annual Journal Metrics
2022 Citation Impact 4.2 - 2-year Impact Factor 4.7 - 5-year Impact Factor 1.131 - SNIP (Source Normalized Impact per Paper) 0.937 - SJR (SCImago Journal Rank)
2023 Speed 19 days submission to first editorial decision for all manuscripts (Median) 135 days submission to accept (Median)
2023 Usage 2,970,572 downloads 1,619 Altmetric mentions
- More about our metrics
- Follow us on Twitter
BMC Microbiology
ISSN: 1471-2180
- General enquiries: [email protected]
Suggestions or feedback?
MIT News | Massachusetts Institute of Technology
- Machine learning
- Social justice
- Black holes
- Classes and programs
Departments
- Aeronautics and Astronautics
- Brain and Cognitive Sciences
- Architecture
- Political Science
- Mechanical Engineering
Centers, Labs, & Programs
- Abdul Latif Jameel Poverty Action Lab (J-PAL)
- Picower Institute for Learning and Memory
- Lincoln Laboratory
- School of Architecture + Planning
- School of Engineering
- School of Humanities, Arts, and Social Sciences
- Sloan School of Management
- School of Science
- MIT Schwarzman College of Computing
Turning microbiome research into a force for health
Press contact :.
Previous image Next image
The microbiome comprises trillions of microorganisms living on and inside each of us. Historically, researchers have only guessed at its role in human health, but in the last decade or so, genetic sequencing techniques have illuminated this galaxy of microorganisms enough to study in detail.
As researchers unravel the complex interplay between our bodies and microbiomes, they are beginning to appreciate the full scope of the field’s potential for treating disease and promoting health.
For instance, the growing list of conditions that correspond with changes in the microbes of our gut includes type 2 diabetes, inflammatory bowel disease, Alzheimer’s disease, and a variety of cancers.
“In almost every disease context that’s been investigated, we’ve found different types of microbial communities, divergent between healthy and sick patients,” says professor of biological engineering Eric Alm. “The promise [of these findings] is that some of those differences are going to be causal, and intervening to change the microbiome is going to help treat some of these diseases.”
Alm’s lab, in conjunction with collaborators at the Broad Institute of MIT and Harvard, did some of the early work characterizing the gut microbiome and showing its relationship to human health. Since then, microbiome research has exploded, pulling in researchers from far-flung fields and setting new discoveries in motion. Startups are now working to develop microbiome-based therapies, and nonprofit organizations have also sprouted up to ensure these basic scientific advances turn into treatments that benefit the maximum number of people.
“The first chapter in this field, and our history, has been validating this modality,” says Mark Smith PhD ’14, a co-founder of OpenBiome, which processes stool donations for hospitals to conduct stool transplants for patients battling gut infection. Smith is also currently CEO of the startup Finch Therapeutics, which is developing microbiome-based treatments. “Until now, it’s been about the promise of the microbiome. Now I feel like we’ve delivered on the first promise. The next step is figuring out how big this gets.”
An interdisciplinary foundation
MIT’s prominent role in microbiome research came, in part, through its leadership in a field that may at first seem unrelated. For decades, MIT has made important contributions to microbial ecology, led by work in the Parsons Laboratory in the Department of Civil and Environmental Engineering and by scientists including Institute Professor Penny Chisholm.
Ecologists who use complex statistical techniques to study the relationships between organisms in different ecosystems are well-equipped to study the behavior of different bacterial strains in the microbiome.
Not that ecologists — or anyone else — initially had much to study involving the human microbiome, which was essentially a black box to researchers well into the 2000s. But the Human Genome Project led to faster, cheaper ways to sequence genes at scale, and a group of researchers including Alm and visiting professor Martin Polz began using those techniques to decode the genomes of environmental bacteria around 2008.
Those techniques were first pointed at the bacteria in the gut microbiome as part of the Human Microbiome Project, which began in 2007 and involved research groups from MIT and the Broad Institute.
Alm first got pulled into microbiome research by the late biological engineering professor David Schauer as part of a research project with Boston Children’s Hospital. It didn’t take much to get up to speed: Alm says the number of papers explicitly referencing the microbiome at the time could be read in an afternoon.
The collaboration, which included Ramnik Xavier, a core institute member of the Broad Institute, led to the first large-scale genome sequencing of the gut microbiome to diagnose inflammatory bowel disease. The research was funded, in part, by the Neil and Anna Rasmussen Family Foundation.
The study offered a glimpse into the microbiome’s diagnostic potential. It also underscored the need to bring together researchers from diverse fields to dig deeper.
Taking an interdisciplinary approach is important because, after next-generation sequencing techniques are applied to the microbiome, a large amount of computational biology and statistical methods are still needed to interpret the resulting data — the microbiome, after all, contains more genes than the human genome. One catalyst for early microbiome collaboration was the Microbiology Graduate PhD Program, which recruited microbiology students to MIT and introduced them to research groups across the Institute.
As microbiology collaborations increased among researchers from different department and labs, Neil Rasmussen, a longtime member of the MIT Corporation and a member of the visiting committees for a number of departments, realized there was still one more component needed to turn microbiome research into a force for human health.
“Neil had the idea to find all the clinical researchers in the [Boston] area studying diseases associated with the microbiome and pair them up with people like [biological engineers, mathematicians, and ecologists] at MIT who might not know anything about inflammatory bowel disease or microbiomes but had the expertise necessary to solve big problems in the field,” Alm says.
In 2014, that insight led the Rasmussen Foundation to support the creation of the Center for Microbiome Informatics and Therapeutics (CMIT), one of the first university-based microbiome research centers in the country. CMIT is based at the MIT Institute for Medical Engineering and Science (IMES).
Tami Lieberman, the Hermann L. F. von Helmholtz Career Development Professor at MIT, whose background is in ecology, says CMIT was a big reason she joined MIT’s faculty in 2018. Lieberman has developed new genomic approaches to study how bacteria mutate in healthy and sick individuals, with a particular focus on the skin microbiome.
Laura Kiessling, a chemist who has been recognized for contributions to our understanding of cell surface interactions, was also quick to join CMIT. Kiessling, the Novartis Professor of Chemistry, has made discoveries relating to microbial mechanisms that influence immune function. Both Lieberman and Kiessling are also members of the Broad Institute.
Today, CMIT, co-directed by Alm and Xavier, facilitates collaborations between researchers and clinicians from hospitals around the country in addition to supporting research groups in the area. That work has led to hundreds of ongoing clinical trials that promise to further elucidate the microbiome’s connection to a broad range of diseases.
Fulfilling the promise of the microbiome
Researchers don’t yet know what specific strains of bacteria can improve the health of people with microbiome-associated diseases. But they do know that fecal matter transplants, which carry the full spectrum of gut bacteria from a healthy donor, can help patients suffering from certain diseases.
The nonprofit organization OpenBiome, founded by a group from MIT including Smith and Alm, launched in 2012 to help expand access to fecal matter transplants by screening donors for stool collection then processing, storing, and shipping samples to hospitals. Today OpenBiome works with more than 1,000 hospitals, and its success in the early days of the field shows that basic microbiome research, when paired with clinical trials like those happening at CMIT, can quickly lead to new treatments.
“You start with a disease, and if there’s a microbiome association, you can start a small trial to see if fecal transplants can help patients right away,” Alm explains. “If that becomes an effective treatment, while you’re rolling it out you can be doing the genomics to figure out how to make it better. So you can translate therapeutics into patients more quickly than when you’re developing small-molecule drugs.”
Another nonprofit project launched out of MIT, the Global Microbiome Conservancy, is collecting stool samples from people living nonindustrialized lifestyles around the world, whose guts have much different bacterial makeups and thus hold potential for advancing our understanding of host-microbiome interactions.
A number of private companies founded by MIT alumni are also trying to harness individual microbes to create new treatments, including, among others, Finch Therapeutics founded by Mark Smith; Concerto Biosciences, co-founded by Jared Kehe PhD ’20 and Bernardo Cervantes PhD ’20; BiomX, founded by Associate Professor Tim Lu; and Synlogic, founded by Lu and Jim Collins, the Termeer Professor of Medical Engineering and Science at MIT.
“There’s an opportunity to more precisely change a microbiome,” explains CMIT’s Lieberman. “But there’s a lot of basic science to do to figure out how to tweak the microbiome in a targeted way. Once we figure out how to do that, the therapeutic potential of the microbiome is quite limitless.”
Share this news article on:
Related links.
- MIT Center for Microbiome Informatics and Therpeutics
- Lieberman Lab
- Broad Institute
- Department of Biological Engineering
- Department of Civil and Environmental Engineering
Related Topics
- Biological engineering
- Innovation and Entrepreneurship (I&E)
- Drug development
- Institute for Medical Engineering and Science (IMES)
Related Articles
![microbiology research studies “The biology [around gut bacteria’s influence on health] is fairly complex, and we’re still in the early days of unravelling it, but there have been a number of clinical studies that have reported benefits to restoring gut health, and that’s our north star: the clinical data,” Finch co-founder and Chief Executive Officer Mark Smith PhD ’14 says.](https://news.mit.edu/sites/default/files/styles/news_article__archive/public/images/202007/MIT-Finch-01.jpg?itok=nMuVS-Q9)
Finch Therapeutics unleashes the power of the gut

A comprehensive catalogue of human digestive tract bacteria

3Q: Eric Alm on the mysteries of the microbiome

Studying the hotbed of horizontal gene transfers

MIT startup tackles nasty infection with first public stool bank
Previous item Next item
More MIT News

A biomedical engineer pivots from human movement to women’s health
Read full story →

MIT tops among single-campus universities in US patents granted

A new way to detect radiation involving cheap ceramics

A crossroads for computing at MIT

Growing our donated organ supply
New AI method captures uncertainty in medical images
- More news on MIT News homepage →
Massachusetts Institute of Technology 77 Massachusetts Avenue, Cambridge, MA, USA
- Map (opens in new window)
- Events (opens in new window)
- People (opens in new window)
- Careers (opens in new window)
- Accessibility
- Social Media Hub
- MIT on Facebook
- MIT on YouTube
- MIT on Instagram
- Search Menu
- FEMS Microbiology Ecology
- FEMS Microbiology Letters
- FEMS Microbiology Reviews
- FEMS Yeast Research
- Pathogens and Disease
- FEMS Microbes
- Awards & Prizes
- Editor's Choice Articles
- Thematic Issues
- Virtual Special Issues
- Call for Papers
- Journal Policies
- Open Access Options
- Submit to the FEMS Journals
- Why Publish with the FEMS Journals
- About the Federation of European Microbiological Societies
- About the FEMS Journals
- Advertising and Corporate Services
- Conference Reports
- Editorial Boards
- Investing in Science
- Journals Career Network
- Journals on Oxford Academic
- Books on Oxford Academic

Six Key Topics in Microbiology: 2024
This collection from the FEMS journals presents the latest high-quality research in six key topic areas of microbiology that have an impact across the world. All of the FEMS journals aim to serve the microbiology community with timely and authoritative research and reviews, and by investing back into the science community .
Interested in publishing your research relevant to the six key microbiology topics?
Learn more about why the FEMS journals are the perfect home for your microbiology research.
Browse the collection categories:
Antimicrobial resistance, environmental microbiology, pathogenicity and virulence, biotechnology and synthetic biology, microbiomes, food microbiology.

FEMS and Open Access: Embracing an Open Future
As of January 2024, FEMS has flipped four of its journals to fully open access (OA), making six out of its seven journals OA. FEMS Microbiology Letters remains a subscription journal and free to publish in.
We are excited to be making high quality science freely available to anyone to read anywhere in the world and further supporting the advancement of our discipline.
View our FAQs page

Never miss the latest research from the FEMS Journals
Stay up to date on the latest microbiology research with content alerts delivered directly to your inbox. This free service from OUP allows you to create custom email alerts to make sure you never miss our on the latest research from your favorite FEMS journals.
Learn more & sign up
Latest posts on X
Affiliations.
- Copyright © 2024
- About Oxford Academic
- Publish journals with us
- University press partners
- What we publish
- New features
- Open access
- Institutional account management
- Rights and permissions
- Get help with access
- Accessibility
- Advertising
- Media enquiries
- Oxford University Press
- Oxford Languages
- University of Oxford
Oxford University Press is a department of the University of Oxford. It furthers the University's objective of excellence in research, scholarship, and education by publishing worldwide
- Copyright © 2024 Oxford University Press
- Cookie settings
- Cookie policy
- Privacy policy
- Legal notice
This Feature Is Available To Subscribers Only
Sign In or Create an Account
This PDF is available to Subscribers Only
For full access to this pdf, sign in to an existing account, or purchase an annual subscription.
An official website of the United States government
The .gov means it's official. Federal government websites often end in .gov or .mil. Before sharing sensitive information, make sure you're on a federal government site.
The site is secure. The https:// ensures that you are connecting to the official website and that any information you provide is encrypted and transmitted securely.
- Publications
- Account settings
- Browse Titles
NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.

Microbiology in the 21st Century: Where Are We and Where Are We Going?
- Copyright and Permissions
- The impact of microbes on the health of the planet and its inhabitants;
- The fundamental significance of microbiology to the study of all life forms;
- Research challenges faced by microbiologists and the barriers to meeting those challenges;
- The need to integrate microbiology into school and university curricula; and
- Public microbial literacy.
This is an exciting time for microbiology. We are becoming increasingly aware that microbes are the basis of the biosphere. They are the ancestors of all living things and the support system for all other forms of life. Paradoxically, certain microbes pose a threat to human health and to the health of plants and animals. As the foundation of the biosphere and major determinants of human health, microbes claim a primary, fundamental role in life on earth. Hence, the study of microbes is pivotal to the study of all living things, and microbiology is essential for the study and understanding of all life on this planet.
Microbiology research is changing rapidly. The field has been impacted by events that shape public perceptions of microbes, such as the emergence of globally significant diseases, threats of bioterrorism, increasing failure of formerly effective antibiotics and therapies to treat microbial diseases, and events that contaminate food on a large scale. Microbial research is taking advantage of the technological advancements that have opened new fields of inquiry, particularly in genomics. Basic areas of biological complexity, such as infectious diseases and the engineering of designer microbes for the benefit of society, are especially ripe areas for significant advancement. Overall, emphasis has increased in recent years on the evolution and ecology of microorganisms. Studies are focusing on the linkages between microbes and their phylogenetic origins and between microbes and their habitats. Increasingly, researchers are striving to join together the results of their work, moving to an integration of biological phenomena at all levels.
While many areas of the microbiological sciences are ripe for exploration, microbiology must overcome a number of technological hurdles before it can fully accomplish its potential. We are at a unique time when the confluence of technological advances and the explosion of knowledge of microbial diversity will enable significant advances in microbiology, and in biology in general, over the next decade. To make the best progress, microbiology must reach across traditional departmental boundaries and integrate the expertise of scientists in other disciplines. Microbiologists are becoming increasingly aware of the need to harness the vast computing power available and apply it to better advantage in research. Current methods for curating research materials and data should be rethought and revamped. Finally, new facilities should be developed to house powerful research equipment and make it available, on a regional basis, to scientists who might otherwise lack access to the expensive tools of modern biology.
It is not enough to accomplish cutting-edge research. We must also educate the children and college students of today, as they will be the researchers of tomorrow. Since microbiology provides exceptional teaching tools and is of pivotal importance to understanding biology, science education in schools should be refocused to include microbiology lessons and lab exercises. At the undergraduate level, a thorough knowledge of microbiology should be made a part of the core curriculum for life science majors.
Since issues that deal with microbes have a direct bearing on the human condition, it is critical that the public-at-large become better grounded in the basics of microbiology. Public literacy campaigns must identify the issues to be conveyed and the best avenues for communicating those messages. Decision-makers at federal, state, local, and community levels should be made more aware of the ways that microbiology impacts human life and the ways school curricula could be improved to include valuable lessons in microbial science.
- Front Matter
The American Academy of Microbiology is the honorific leadership group of the American Society for Microbiology. The mission of the American Academy of Microbiology is to recognize scientific excellence and foster knowledge and understanding in the microbiological sciences.
The opinions expressed in this report are those solely of the colloquium participants and may not necessarily reflect the official position of the American Society for Microbiology.
Board of Governors, American Academy of Microbiology
Eugene W. Nester, Ph.D. (Chair) University of Washington
Kenneth I. Berns, M.D., Ph.D. University of Florida Genetics Institute
James E. Dahlberg, Ph.D. University of Wisconsin, Madison
Arnold L. Demain, Ph.D. Drew University
E. Peter Greenberg, Ph.D. University of Iowa
J. Michael Miller, Ph.D. Centers for Disease Control and Prevention
Stephen A. Morse, Ph.D. Centers for Disease Control and Prevention
Harriet L. Robinson, Ph.D. Emory University
Abraham L. Sonenshein, Ph.D. Tufts University Medical Center
David A. Stahl, Ph.D. University of Washington
Judy D. Wall, Ph.D. University of Missouri
Colloquium Steering Committee
Roberto G. Kolter, Ph.D. Harvard Medical School (Co-Chair)
Moselio Schaechter, Ph.D. San Diego State University (Co-Chair)
Stanley R. Maloy, Ph.D. San Diego State University
David A. Relman, M.D. Stanford University School of Medicine
Margaret A. Riley, Ph.D. Yale University
Carol A. Colgan, Director American Academy of Microbiology
Colloquium Participants
Victor de Lorenzo, Ph.D. Centro Nacional de Biotecnologia, Madrid, Spain
William E. Goldman, Ph.D. Washington University of Medicine, St. Louis
Peter M. Hecht, Ph.D. Microbia, Inc., Cambridge, Massachusetts
Laura A. Katz, Ph.D. Smith College
Roberto G. Kolter, Ph.D. Harvard Medical School
Mary E. Lidstrom, Ph.D. University of Washington
William W. Metcalf, Ph.D. University of Illinois
Eugene W. Nester, Ph.D. University of Washington
Gary J. Olsen, Ph.D. University of Illinois
Moselio Schaechter, Ph.D. San Diego State University
Gisela Storz, Ph.D. National Institutes of Health
Saeed Tavazoie, Ph.D. Princeton University
Jennifer J. Wernegreen, Ph.D. Marine Biology Laboratory, Woods Hole, Massachusetts
Merry Buckley, Ph.D. Freelance Science Writer, Ithaca, New York
- Executive Summary
- Introduction
Microbiology has never been more exciting or important than it is today. Powerful new technologies, including novel imaging techniques, genomics, proteomics, nanotechnology, rapid DNA sequencing, and massive computational capabilities have converged to make it possible for scientists to delve into inquiries that many thought would never be approachable. As a result, hardly a day goes by without another discovery that points to the central importance microbial life has in carrying out the cycles of gases and nutrients that sustain all life and affect conditions on this planet. The increasing human population, combined with increases in global travel, has apparently created a sharp rise in the emergence and re-emergence of infectious diseases, alarming the public and frustrating public health officials.
Issues of microbial contamination are also more pressing now than ever. The microbial quality of our food and water in a crowded, complex world must be vigorously addressed to maintain health and a high quality of life for the all citizens of the world. Finally, a bioterrorism event involving spores of Bacillis anthracis occurred in the United States in 2001, and continuing investigations worldwide reveal that bioterrorism is a genuine threat from ill-intentioned groups and individuals using other microbes and toxins.
As microbiology is faced with this tumult of advancements, opportunities, problems, and threats, the science stands at the threshold of a new era. But in what direction is microbial science going? What is really important and what is merely distraction? What will best improve people's lives and the health of our shared planet? In short, what new directions should microbiology take in the 21 st century?
Microbiologists want to know how microbial science is changing in the wake of advancements in technology and growing human pressures on the world's resources. They want to know what topics deserve exploration and where the obstacles to exploring those areas lie. As we stand at the convergence of genomics, public concerns about bioterrorism, global outbreaks of infectious diseases, unprecedented computational power, and the possibility of large-scale ecological disasters, where do the greatest opportunities lie in microbiology and what obstacles must be overcome for these opportunities to be realized?
It is clear to microbiologists that microbes are the basis of the biosphere; they are the support system for life on earth and the wellspring from which all other life has arisen through billions of years of evolution. Since microbes are fundamental to life and microbial science is in the headlines now more than ever before, it follows that educating young people in microbial science is critical. But where does microbiology fit into existing school curricula? How can college biology coursework be updated to reflect more accurately the pivotal place of microbial science in understanding our world? How can the public-at-large be made aware of microbes, the role of microbes in sustaining life, and the danger posed by modern infectious diseases? And how should public outreach campaigns be conducted? What are the important messages to deliver? How can the importance of microbes and microbiology be conveyed to people in power, the decision-makers?
To answer such questions, the American Academy of Microbiology convened a colloquium in Charleston, South Carolina on September 5–7, 2003. Experts in the fields of bacteriology, virology, eukaryotic microbiology, medicine, biotechnology, molecular biology, and education met to discuss the central role of microbes in maintaining life on earth, the current research challenges that face the field, the pivotal role of microbiology education to training in all life sciences, and methods for encouraging public literacy of microbial science.
- The Central Role of Microbes and Microbiology
Microbes affect all life and the physical and chemical make-up of our planet. They have done so since the origin of life. No other group of organisms can make such a claim. Life without all other creatures is possible, but life without microbes is not. Consequently, we believe that one cannot carry out in-depth studies of any branch of biology or geology without taking into account the activities of microbes. Microbes are the masters of the biosphere, and ours indeed is a planet of the microbes.
Microorganisms are also determinants of human health and the source of critical materials for medical and industrial use. Microbiology, therefore, is as central to the study of life as biochemistry, genetics, evolution, or molecular biology. The informed biologist must treat microbiology as core and not as a particular branch of biology.
Microbes As the Basis of Life
The root of the tree of life.
Microbes were the progenitors of all the complex and varied biological forms that now exist on Earth. Plants and animals emerged within a microbial world and have retained intimate connections with, and dependency upon, microorganisms. As the root of the tree of life, microbes were the original templates from which all life was formed and to which all life has an intimate familiarity. In order to understand the evolution of organisms we see today at the tips of the branches of the tree of life, it is necessary to study how they are related to their ancestors and what those ancestors were like.
By studying the microorganisms living today that echo the properties of the first life forms, microbiologists seek to understand the forces and processes that created our global ecosystem. Moreover, microorganisms are the preeminent systems to use in experimental evolution, as they offer the researcher fast generation times, genetic flexibility, unequaled experimental scale, and manageable study systems. Studies using microbes have led to groundbreaking insights into the evolution of all species. For example, investigations of the interrelatedness of microbes first brought to light the current model of the evolutionary relatedness of all life on earth, the tree of life (See Box 1 – the Tree of Life).
The Tree of Life Before and After Molecular Microbiology.
MAINTAINING LIFE ON EARTH
Life not only began with microorganisms, the continued existence of life on earth totally relies on the inconspicuous microbe. It has been estimated that a staggering 5×10 31 (50,000,000,000,000,000,000,000,000,000,000—weighing more than 50 quadrillion metric tons) microbial cells exist on this planet, and it is difficult to overstate their importance to the biosphere. Microbes are responsible for cycling the critical elements for life, including carbon, nitrogen, sulfur, hydrogen, and oxygen. By cycling these elements in soils, microbes regulate the availability of plant nutrients, thereby governing soil fertility and enabling the efficient plant growth that sustains human and animal life. Microbes also play a big role in cycling atmospheric gases, including the compounds responsible for the “greenhouse effect,” which, paradoxically, sustains life on our planet, but through global warming, poses a threat to all living things. More photosynthesis is carried out by microbes than by green plants. It turns out that, excluding cellulose, microbes constitute approximately 90% of the biomass of the whole biosphere (more than 60% if cellulose is considered).
Since microbes can take up nutrients and other elements that larger organisms often cannot exploit, microorganisms are positioned at the base of many food chains, where they siphon previously inert, inorganic materials into the biosphere. Microbes are also master recycling experts; they degrade biological wastes and release the critical elements for use by other organisms.
Scientists have only begun to understand the ways that microorganisms are tuned into their environments, how they respond to changes, and how they communicate with other members of microbial communities to carry out the functions that sustain the biosphere. Understanding these phenomena will lead to a more complete knowledge of our global ecosystem and may allow scientists to correct human damage to ecosystems, large and small. Humans are latecomers to this planet, and a great deal may be learned from microbes about the maintenance of essential planetary processes.

VITAL BUT DANGEROUS TO HUMAN HEALTH
Humans have an intimate relationship with microorganisms. Despite their overwhelmingly beneficial impact on the environment, a small notorious set of bacteria, fungi, parasites, and viruses may cause disease. In the struggle against disease, our bodies attempt to establish a delicate balance between the microorganisms and viruses that are beneficial to our health and those that exploit the human host to the body's detriment. More than 90% of the cells in our bodies are microorganisms; bacteria and fungi populate our skin, mouth, and other orifices. Microbes enable efficient digestion in our guts, synthesize essential nutrients, and maintain benign or even beneficial relationships with the body's organs. The presence of these organisms influences our physical and mental health. In experiments, it has been found that sterile animals are markedly less healthy than animals that have been naturally colonized by microorganisms.
How do microorganisms cause disease? Pathogenic microorganisms and viruses have an individual ecological strategy that determines where they strike and what impacts they have on the host. One root of the problem is that pathogens colonize areas within the human body that our immune system sees as “privileged.” In the process of gaining access to these locations or in maintaining their colonies, microorganisms and viruses may cause damage to human tissues, creating signs and symptoms of disease. Disease may also begin when the immune system detects a microbial cell or virus. The body's immune system responds with an attack on the foreign organism that may cause harm to the body itself. In some cases, the damage caused by pathogens in human tissues or the immune response to them can promote the transmission of the pathogen to a new host. Hence, causing disease is just another ecological strategy for certain microorganisms—one in which the human body is used as a habitat for multiplication, persistence, and transmission.
Disease emergence—the situation where a new disease-causing microbe or virus is identified or an old one causes a new disease—is a hot-button topic today. From the E. coli strain O157 to SARS, new diseases and new pathogens are identified every year, frightening the public and confounding public health officials responsible for stemming the tide of outbreaks. Many circumstances likely play a role in the increased rate of disease emergence, including a variety of host, environmental, and social factors.
USEFUL TO INDUSTRY AND MEDICINE
Industry and medicine are increasingly reliant on microorganisms to generate chemicals, antibiotics, and enzymes that improve our world and save lives. Microbes are being domesticated with the tools of molecular biology for production of biodegradable plastics and all types of new materials. Biotechnology, which will soon be a pillar of the industrial base in the U.S., employs microorganisms and viruses in a number of ways, including such divergent applications as the genetic engineering of crops and gene therapy. Microbiology research has enabled these successful technologies, and future advancements in using microbes in industry and medicine rely on conducting effective research today.
- Microbiology as the Foundation of Biology
In light of the critical functions microbes carry out to the benefit and detriment of life on earth, the study of microbiology must be treated as a core subject. Moreover, microbes are ideal experimental systems for investigating many of the otherwise confounding key questions in biology.
Microbiology directly provides important tools for experimental science. Because of their relative simplicity, microbes are ideal systems for sorting out basic questions about the origin of sex, speciation, adaptation, cellular function, genetics, biochemistry, and physical properties of all other living organisms. Of particular significance is the ability, using single-celled microorganisms, to match a gene with a characteristic of the organism, otherwise known as linking genotype with phenotype. Microbial cells in culture are not the only available microbial tools. Microbial communities can be put to good use in exploring ecological principles and identifying the metabolic properties, interactions, and communications at work in a relatively simple ecosystem.
The “virtual microbial cell” (a complete computer simulation of the minimal set of genes and functions at work in one live bacterium or yeast) also may be highly instructive, allowing researchers to build models of complete metabolic pathways, cell circuits, and other phenomena to create a virtual network that describes a cell. Such a virtual status may, in fact, soon turn into a physical reality, as the emerging field of Synthetic Biology (building up bacteria from scratch endowed with desired properties) develops and comes to fruition.
Current Issues and Research Challenges
- “Calm urged over mystery virus; a flu-like illness has downed at least 50 staff in two days at the Prince of Wales Hospital,” South China Morning Post (Hong Kong), March 13, 2003.
- “Probe Begins Trip to Mars In Quest for Water, and Life,” The New York Times, June 11, 2003.
- “3rd Death in Hepatitis Outbreak; Pa. probe focuses on handling of produce,” Newsday, November 15, 2003.
- “Engineered corn found to kill butterflies,” Milwaukee Journal Sentinel, August 22, 2000.
- “Infections eyed as cause of cancer, heart disease,” The Boston Herald, April 18, 1999.
Today, microbiology is in the headlines more than ever before, and research behind the headlines, and behind other critical issues of which the public is largely unaware, is changing rapidly.
HOW THE IMPORTANCE OF MICROBIOLOGY TO SOCIETY HAS CHANGED IN THE LAST TEN YEARS
The public is now more aware of microorganisms and viruses than at any other time in history. Unfortunately, that public awareness is usually laced with anxiety and dread. Events of the last ten years and the tone of the media coverage of those events have served to feed the public's fear and create the perception of an increased risk from the microbial world. Such “microbiophobia” has resulted in surges in the popularity of disinfectants, antimicrobial soaps, and other products that purport to keep disease at bay. Bioterrorism and the distinct possibility that anthrax or another infectious microbe could be used as a weapon against innocent civilians, crops, or livestock have frightened people across the globe. An apparent increase in the emergence of novel infectious diseases, including SARS, West Nile disease, and others, has also brought microbes into the public eye. Recently, conventional health therapies to combat certain infectious diseases, including AIDS and tuberculosis, have failed due to the ongoing evolution of these pathogens, heightening public doubts about the ability of scientists and physicians to protect the public even from familiar diseases. A few chronic diseases that were once thought to be due to factors like genetic susceptibility or chance have instead been shown to be the work of bacteria or viruses. And in the U.S. and other developed nations, large-scale food contamination events are on the rise, often sickening tens or hundreds of people before public health officials can identify the sources of infection and restrict the public's exposure.
Advancements in microbiology over the last ten years are frequently overlooked in the wake of public concerns about biowarfare, infectious disease, and foodborne illness. Yet, the progress of the last decade is undeniable. Pharmaceutical research now relies heavily on microbes and microbiology for drug discovery and production. Green chemistry, in which microorganisms are employed to carry out industrial processes, is an increasingly effective strategy for tackling issues of safety and sustainability in chemical-related industries. Biotechnology, too, relies on microbial technologies and microbial genes for carrying out modifications that improve crops, breeds of livestock, and synthetic feed-stocks. In agriculture, microbes and microbial products are now used in probiotic therapies, antibiotics, and pest control measures. Advancements in food microbiology have improved the safety of the food we buy in our supermarkets and restaurants, doubtlessly saving lives every single day. At hazardous waste sites, microbes have been put to work digesting noxious chemicals—metabolizing them into harmless materials, thereby preventing further contamination of soil and water. Bioterrorism and disease are frightening, but progress in microbiology and advancements in applying microbes to solve seemingly intractable human problems should be kept in mind.
How Research Is Changing
Not only has the public's perception of microbiology changed in the last decade, the practice of microbiology research has been altered as well. The past ten years of microbiology have been dynamic and exciting, and new discoveries have been built upon the remarkable work of the past.
MORE SYNTHESIS, LESS REDUCTIONISM
Microbiology once focused almost solely on individual microorganisms grown in isolation under artificial conditions, attempting to extrapolate an understanding of disease or the environment from minute observations recorded in the laboratory. Today, however, much of the science is moving away from reductionist approaches and into the realm of synthesis—weaving together a fabric of measurements and observations of the microorganism, its environment, and the influence of other organisms at many scales to create an integrative picture of microbial activities. Once unfamiliar, the concept that cells consist of a network of interacting proteins now permeates the science. A greater emphasis is now being placed on systems-level research, in which microbes in their habitats are being treated as a series of interrelated compartments, processes, and feedbacks. Placing a synthetic or systems lens on microbiology can be highly instructive and has several advantages over strict reductionism. It is hoped that in the future, integrative approaches will enable microbiologists to predict microbiological outcomes, allowing them to pinpoint the consequences of a perturbation of human health or of a given ecosystem.
INCREASED EMPHASIS ON EVOLUTION AND ECOLOGY
In the past ten years, microbiologists have increasingly recognized the importance of ecology and evolution. Studies in experimental ecology and evolution have provided evidence on the principles that apply not only to microbes, but possibly to larger organisms as well. Ecological thinking has become dominant, and microbiology is no longer the test tube science of the past. An example is the realization that the way microbes cause disease is, in fact, an ecological problem requiring understanding of both the microbe and its environment—the host in the case of disease.
A change has also taken place in the investigative style of research in microbiology. Previously, many lines of inquiry were closed due to technological limitations or a lack of expertise in fields tangential to microbiology, like soil science, geology, or medicine. In these cases, the problem under investigation was often re-defined to suit the techniques at hand and the academic experience of the principal investigator. Today, these lines of inquiry are often explored head-on by applying the technology advancements of the last ten years and, more importantly, recruiting expertise and resources from other disciplines, often through collaborations.
TECHNOLOGICAL ADVANCEMENTS
Technological progress has reformed the landscape of microbiology research, making long-standing questions about microbes finally amenable to study. Chief among the significant advancements of the last ten years is the development of technologies that make genomics possible, including increased computational power, more rapid DNA sequencing, and other laboratory techniques. Genomics employs all or part of the genome, the full genetic complement of a cell, to answer questions about an organism. Although genomics has impacted most of the life sciences and enabled new insights into the functions and processes of all life forms, its most significant impact has been on microbiology, a development that has opened new insights into the ecology and evolution of microorganisms. Other large-scale research, such as proteomics or transcriptomics (the pattern of gene expression), has also had a great impact on the practice of microbiology research. Improvements in information technology have increased interactions between researchers of all fields, enabling a continuing dialogue on the commonalties between microbiology and other disciplines. Nanotechnology and related approaches should allow researchers to experiment with single cells, answering long-standing questions about microbial physiology. Finally, high-end imaging techniques such as nuclear magnetic resonance imaging (NMR), ESR, and others have allowed detailed analyses of microbial cell structure and the structure of microbial communities.
With the advent of molecular microbiology, traditional approaches for defining microbial causation of disease, such as Koch's postulates, have been found insufficient, as they oftentimes lead to “false negative” conclusions. Researchers have struggled with creating robust standards for identifying microbial causation that go beyond Koch's postulates and make use of technological advancements to identify causative links even for microbes that cannot be cultivated in the lab.
- Hot Research Topics
A number of areas of microbiology research are particularly topical in the wake of technological advancements and discoveries that have brought to light previously unexplored aspects of microbial life. Topics including genomics, biocomplexity, infectious disease, the origins of life, and the application of microbes to improve quality of life are at the forefront of the list of previously unattainable research areas that are being actively pursued today.
ENVIRONMENTAL GENOMICS AND ENVIRONMENTAL METAGENOMICS
Bacteria and archaea tend to have smaller genomes than eukarytic cells, which makes them more amenable to sequencing. The study of genomics has had a huge impact on microbiology. Lines of inquiry related to the factors that govern microbial genome organization, dynamics, and stability are highly approachable using these genomic techniques. But, despite the vast tracts of sequence data that are available, more rapid and accurate methods of annotation attributing a function to a gene are sorely needed. Scientists can now explore questions related to the extents of diversity within naturally occurring microbial communities and to the functional significance of that diversity. Metagenomic technologies are being used to examine the DNA of nonculturable bacteria and microbial consortia without any sub-culturing, thereby allowing us to understand the interplay of genes and functions in an ecosystem, regardless of the specific microbial hosts. We can now ask how the genes of all members of a community relate to the functions carried out by that particular community.
Many other critical questions about microbial life may now be addressed using genomics. Interested readers are referred to the American Academy of Microbiology's colloquia reports “The Global Genome Question: Microbes as the Key to Understanding Evolution and Ecology” and “Microbial Ecology and Genomics: A Crossroads of Opportunity” (see http://www.asm.org ).
BIOCOMPLEXITY
In addition to genomics-related topics, questions related to biocomplexity are at the forefront of microbiology research. Biocomplexity in microbiology encompasses the interactions among microbes and between microbes and their environment. The emphasis in biocomplexity research is on the whole ecosystem, rather than its parts, seeking to identify the emergent properties that cannot be found in studies of individual components. Interdisciplinary collaboration is inherent in this kind of research since it often calls for the expertise of environmental engineers, biologists who study larger organisms, soil scientists, hydrologists, marine biologists, and other related professionals. Research in biocomplexity should progress rapidly in the coming years. Questions on the shape of microbial biocomplexity, its temporal and spatial variability, will doubtless be investigated. Other questions related to microbial biocomplexity are the definition of a microbial “species,” how species are created, and at what rate.
INFECTIOUS DISEASE
Grappling with topics related to infectious disease is certainly not new for microbiologists, but the discoveries and advancements of the past ten years have revealed new horizons in the field, presenting exciting opportunities to improve the quality of human life. The ability to predict the emergence of disease is a particularly critical topic. Research into the environmental factors that trigger the emergence of pathogens, the factors that drive disease migration, and seasonal patterns in disease frequency may shed light on the factors that affect how new and old diseases emerge and persist in populations. These observations will enable us to design better therapeutic strategies for new and existing pathogens.
Recent discoveries that have linked human diseases (e.g., stomach ulcers and cervical cancer) to bacterial or viral causes highlight the possibility that other chronic illnesses with mysterious etiologies may also be microbially mediated. Candidates include inflammatory bowel disease, diabetes, rheumatoid arthritis, sarcoidosis, systemic lupus erythematosus, and coronary artery disease. Research into the causes of these diseases and others will shed more light on these diseases and their diagnosis, prevention, and treatment.
THE ORIGINS AND HISTORY OF LIFE
Science now has better tools at its disposal to explore the origins of life, and microbes are well suited to experimental approaches for understanding these first organisms. The evolutionary origins of sex may also be explored using microbial systems. Analysis of the distribution of sex (here referring to the fusion of gametes) on the emerging tree of life indicates that this process arose very early in the evolution of eukaryotic cells. Research will also focus on assembling the complete tree of life, a comprehensive phylogenetic framework that includes all life forms on earth.
ENGINEERING MICROBES TO IMPROVE THE QUALITY OF LIFE
To an ever-greater extent, microbes can be put to use to improve the human condition. Methods of detecting and identifying novel microbial products are likely to be scrutinized and improved upon, expanding the ability to exploit the metabolic versatility of microbes in providing powerful antibiotics, therapeutics, and other materials. It is also likely that microbes can be put to work in energy recovery and utilization. In this respect, microbial production of H 2 is bound to be one of the keys for addressing the unavoidable shortage of energy in the future and for mitigating the greenhouse effect of fuel combustion.
Other Hot Topics in Microbiology.
- Meeting Future Research Challenges
The future is bright for microbiology. Advancements in the study of infectious disease, microbial ecology, plant and animal pathology, and biotechnology promise to improve human life and the well being of the environment, and new opportunities have come about through social and scientific changes. Progress on these synthetic activities will be hastened through improvements in technology and through changes in education and training.
Technological Hurdles
Several technological hurdles stand before today's microbiology researchers. To make progress, science should not accept the limitations placed on discovery by traditional methods, conventional approaches, or existing infrastructure. Particular attention should be focused on the technologies that enable genomics, single-cell analyses, microbial cultivation, and establishment and maintenance of microbiological databases.
Although progress in microbial genomics is being made at a fantastic rate, availability of appropriate tools still places limits on research. It would be ideal to have the complete genomes of many thousands of species and strains of microbes, but this is currently not possible, given the limits on the speed of sequencing and computational capacity for data manipulation, which both translate into limitations in funds available for such an endeavor. Accelerated and inexpensive sequencing capabilities are needed to conduct sequencing on this scale. In order to interpret microbial genome sequence data in a meaningful way, more tools and approaches beyond those that solely rely on gene homology for inferring gene function are sorely needed. Annotation of genomes is currently a major hurdle for the field, and standards and methods are needed that can accelerate the process and provide consistent high quality results.
STUDYING SINGLE CELLS
The ability to analyze single cells has eluded microbiologists in the past. In order to better understand the activities of microbes in their natural settings, technologies and assays that would allow the monitoring of single cells in a variety of conditions, including in situ , are necessary. Specific capabilities should include genome sequencing, gene expression analysis, and the ability to measure intracellular pools of small molecules. Ideally, these analyses should be amenable to high-throughput approaches.
CULTIVATION
Improved technologies for cultivating diverse microbes are badly needed. It is never far from a microbiologist's mind that more than 99% of microbes have never been cultivated in the laboratory. The fact that the vast majority of microbial life cannot be scrutinized with respect to growth, metabolism, and reproduction comprises a massive gap in our understanding of the microbial world.
Currently, a need exists for quantitative digital formatting of microbiological data in a portable and standardized fashion. To better integrate microbiological data from multiple studies and from multiple laboratories, an effort should be made to standardize data collection and annotation.
Scientific Needs
- Researchers must integrate their work with that of scientists in related fields.
- Computational scientists should become more familiar with and integral to microbiology.
- Microbiology materials and data must be more carefully curated.
- Powerful, but expensive, modern equipment should be housed in community facilities, open to researchers who might not otherwise have access to these technologies.
INTEGRATING DISCIPLINES
The issues surrounding microbiology touch on so many other disciplines that meeting the grand challenges in microbiology requires integrating the expertise of professionals in many fields. The response to public concerns about bioterrorism, for example, presents a formidable task that requires the contributions of micro-biologists, physicians, pathologists, forensic scientists, and others.
In light of the opportunities and challenges in microbiology today, a number of fields of expertise are especially ripe for integration. Pathogenic microbiologists should see themselves as microbial ecologists who should study both the microbe and the host with analogous intensity. Enhancing the linkages between organic chemistry and microbiology would prove helpful to a number of areas of inquiry, including bioremediation and green chemistry. Microbiology should borrow expertise from systems engineering in efforts to create networks of metabolic pathways. Other interdisciplinary opportunities include collaborations with professionals in imaging sciences, statistics, nanotechnology, biosystematics, mathematics, biochemistry, ecology, and structural chemistry. Moreover, some relatively neglected fields within microbiology should be revived and facilitated by integrating with these related disciplines, including microbial physiology and the biology of eukaryotic microbes (fungi, protists). Collaborations between micro-biologists who work with prokaryotic or eukaryotic microbes and virologists should also be encouraged.
Some successful integrations have already taken place. The fields of geology and microbiology have already been joined on a number of levels to cope with questions surrounding the significance of microorganisms in global geological processes, and molecular biology has met up with information science to provide bioinformatics, which is used to manage genetic and protein sequence data.
A number of routes could be developed to foster these integrations. Visiting fellowships could be established to bring professionals with expertise in statistics, biochemistry, or ecology, for example, into microbiology labs and vice versa, placing microbiologists into statistics, biochemistry, or ecology labs. Microbiologists and professionals in related disciplines could also assemble into working communities across departmental boundaries to cooperate on subjects best addressed through multidisciplinary collaborations. Other integrations could be encouraged by funding agencies.
COMPUTATIONAL SCIENCE
There is a need to bring computational science into closer contact with the daily work of microbiology. The basic skills involved in computer science, including programming, for instance, should be acquired, or at least be highly familiar, to the average microbiologist.
CURATION OF MATERIALS AND DATA
A great need exists to improve the current modes of curation, entry, storage, and distribution of materials and data related to microbiology. The procedures surrounding culture collections, in particular, need to be revamped. Distribution of cultures has to be conducted in a way that both respects the need for national security and recognizes the ability of these materials, in the hands of researchers, to further the science that directly benefits society. If the international microbiological community does not confront the need for thoughtful review of potentially problematic materials and data, then mechanisms governing release and distribution of data will be imposed by others.
Progress in microbiology has always been enabled by the technology available, a fact that is still true today. However, many researchers are stymied by a lack of access to the expensive instruments that would enable them to make the greatest strides. Facilities for housing and making these technologies available to microbiology researchers would allow investigators in moderately funded and underfunded labs to achieve their full potential. In these technology centers, investigators could come to conduct work, using techniques like NMR, spectroscopy, and other imaging methods, under the guidance of trained staff. Regional centers could even promote technology development and could play a part in advancing training, education, and out-reach among participating educational institutions.
- Key Opportunities for Microbial Biologists
There are more opportunities available for microbiologists today than at any time in the history of the field. Although the microbiological advancements of the last two centuries have been profound, a great deal of biology remains to be discovered and described through study of the microbial world. Microbiology can be used to push back the frontiers of biology, opening up new ways to harness the power of biology to improve human health and the environment. Microbiologists must participate in this effort.
CAREER OPPORTUNITIES
Career opportunities for microbiologists abound in the wake of new technologies that have changed the face of biology. Biotechnology, in particular, is intimately connected with microbiology and calls for the skills of microbiologists to execute the work that holds the potential to improve the quality of human life. Without a profound grasp of microbiology, much of biotechnology is not possible. As a future pillar of the industrial base in the United States, biotechnology offers many chances for microbiologists to contribute in substantive ways to the future of the world.
Antibiotic discovery is also closely tied to the skills of microbiologists. The importance of this field cannot be overstated, since most individuals in developed countries have experienced first-hand the life-saving power of antibiotic therapies. However, the threat of microbial resistance to antibiotics looms large.
Scientific discoveries can be put into action more rapidly through greater collaborations between academia and industry. By cooperating to develop concepts and inquiries, microbiologists and industrial decision-makers can bring technologies to market or apply microbial solutions to persistent manufacturing problems. Efforts should be made to overcome regulatory and cultural obstacles that stand in the way of such collaborations.
Finally, increased emphasis on systems-level and quantitative research in microbiology has opened new doors for microbiologists working in interdisciplinary research teams or who have backgrounds in other disciplines. Individuals with experience in physics, mathematics, engineering, or computer sciences are in high demand in microbiology today, and this will likely continue for the foreseeable future.
INDUSTRIAL SUSTAINABILITY
As planet resources become more scarce, and environmental awareness is translated into a widespread social demand, industry is bound to reformulate many of its traditional chemically-catalyzed processes into more environmentally-friendly alternatives and products. Every prospective study (for instance the OECD reports “Biotechnology for a Clean Environment” and “The Application of Biotechnology to Industrial Sustainability”) predicts the booming of a new multi-billion dollar market around processes and goods originating in biocatalysis, both for biosynthesis of added-value molecules or for biodegradation and pollutant removal.
The emerging interfaces between chemical engineering and microbial genetics/metabolism will create countless job opportunities for those who seize the right training early enough in the process. The fields of large-scale mining and metallurgy, so far limited to hard-core engineering, will soon benefit from ongoing advances in geomicrobiology, and experts in this field soon will be in great demand. The relatively new field of green chemistry will, thus, offer employment perspectives for microbiologists and present a chance for scientists to work at the forefront of developing sustainable technologies.
NATIONAL DEFENSE
Growing concern about biological security promises to create a number of employment and research opportunities for microbiologists. Bioterrorism has defined a need, in this country and elsewhere, for new and improved infrastructures to address issues related to national security. Microbial science is key to proper execution of these new security measures. The opportunities are diverse; establishment of research centers related to bioterrorism, development of secure culture collections, vaccine development, database development, and other activities will all require the contributions of microbiologists. It is important to note that, with respect to biological security, global preparation requires global knowledge. It is critical for science to protect the freedom to exchange information on the biological agents of disease.
- Training to Meet the Needs and Challenges of the Future
The training of tomorrow's microbiologists is taking place in fourth grade classrooms, in high school biology labs, and in the lecture halls of universities all over the world. Although the educational systems of past and present have produced the great minds of microbiology, improvements need to be made if microbiology is to fulfill its potential in the new century.
Given the central importance of microbial science to biology in general, teaching of microbiology should be thoroughly integrated into school curricula. At the undergraduate level, emphasis needs to be placed on textbook revision and on integrating microbial sciences into the basic coursework for biology.
Microbiology Education in Schools
As both the root of the tree of life and the matrix that supports the biosphere, microbes should take center stage in science curricula at the elementary, middle, and high school levels. If we are to achieve a well-educated public, versed in the fundamentals of biology and capable of tackling the demands of the new century, the importance of microbiology must be acknowledged by teachers and policy-makers and translated into meaningful school lessons. In practice, this means that microbiology should be integrated into all phases of biology education, not segregated as separate coursework or, as is often the case, as a few sessions at the beginning of a biology course. Achieving integration of microbiology in school curricula will require that educational decision-makers understand and acknowledge the magnitude of microbial contributions to life on earth.
Reorienting school curricula begins with changes in biology textbooks. General biology texts should be organized around a microbiology core. In this way, studying micro-biology can enrich the study of plants, insects, and animals. For example, explaining the importance of microbial gut flora to termites would lend depth and greater applicability to the simple lesson that “termites eat wood.” The food chain in the ocean does not start out by small fish being eaten by big fish, but by microbial populations providing the bulk of the organic material required to set the chain in motion. The oxygen we breathe is not made just by plant photosynthesis, but, to an even greater extent, by the activities of microbes.
More specialized books can also be developed to address the “Grand Challenge” questions, those issues that continue to inspire and confound biologists. Such texts can serve to illustrate the latest discoveries, technologies, and the future of inquiry in microbiology.
Changing the textbooks that schools use has, in the past, proven to be an arduous, protracted process. But, educating the public, beginning with young people, about the importance of microbiology in day-to-day life and in the future of industry is more than a worthy goal—it is an imperative.
Games could also be used for injecting microbiology into curricula. Through creative games or video games based on microbial themes like natural selection, teachers can bring the lessons and fascination of microbiology to students in a friendly, hands-on way. Biology education can be made more engaging with microbial demonstrations and hands-on microbiology lab exercises, which are inexpensive and accessible to a wide range of classroom budgets. Centering lab experiments around simple illustrations of microbial phenomena like decomposition or growth would circumvent both the tedium associated with rote memorization of science lessons and the “gross-out factor” involved, e.g., with frog dissection. Placing a microscope and a sleeve of Petri dishes in every classroom would go a long way toward engaging students in microbiology and in the scientific exploration of the world around them. Some of these activities have already been developed, and more should be created.
Better visual aids are also needed in science classrooms; children would find micrographs of elegant and grotesque microbes appealing, for example. One successful demonstration of the power of microbial illustrations in education can be found on the website for the Marine Biological Laboratory at http://microscope.mbl.edu . A powerful resource for teachers is http://microbeworld.org , sponsored, in part, by the American Society for Microbiology.
In high school biology, in particular, microbiology needs to be taught in an appealing, captivating manner. Many current teachers need to be retrained in the technology and theory associated with the modern microbial science.
Training at the Undergraduate Level
At the undergraduate level, microbiology education takes on two different aspects: training future microbiologists and training biologists in other fields. With respect to training the microbiologists of tomorrow, efforts need to be directed toward revising textbooks to reflect new knowledge on the global importance of microbes and toward overcoming the emphasis on memorization that may still plague some microbiology coursework.
It is clear that all life scientists should receive microbiology training as part of their core curriculum. The topics of microbial physiology, evolution, biochemistry, and genetics should all be worked into the curriculum of undergraduate life sciences students. Luckily, there are many opportunities to introduce appropriate microbiology coursework into the curricula of other disciplines. Organic chemistry courses, for example, which are required for almost all biology students, would benefit from examples taken from microbiology and green chemistry to demonstrate the synthesis of complex compounds from simple precursors. Even students in fields outside of the life sciences would benefit from lessons in microbiology, perhaps presented in biology exploration courses for non-majors as a “microbes and you” segment.
In addition to changes in curricula, improvements are needed at the departmental and college levels as well. In many universities, microbiology is treated strictly as a field of specialization, not as a core subject. Given the fundamental significance of microbial sciences, there should be recognition of the importance of a having a critical mass of microbial sciences faculty. Such faculty need not necessarily be housed in microbiology departments. Appointments of microbiologists are highly desirable in departments of geology, chemistry, clinical medicine, engineering, and even history. These faculty can cross traditional departmental barriers to interact across many fields, effectively educating and training the next generation of scientists.
Promoting Microbial Literacy
Review the facts: microbes were the first life forms, they are important determinants of human health, and they carry out the processes that ensure clean drinking water and fertile soil. They are the most genetically and biochemically diverse forms of life and are the most rapidly evolving organisms on the planet. Microbes govern environmental cycling of the world's nutrients and the substances necessary for life. In every crevice and on every surface, from the deep earth's crust to steaming sulfurous plumes, to the gut of every insect on the planet, microbes are there. They are a key component of all biological systems. In light of these truths, it is readily apparent that microbiologists must make an effort to educate both the public and policy-makers. However, it is less obvious which messages should be conveyed and how best to communicate these facts.
Public Literacy
There is a serious gulf between the excitement experienced by those working in microbiology and the level of awareness in the general public. However, there is evidence to indicate that increased levels of support for public literacy on major public health issues like HIV-AIDS, West Nile disease, and SARS, influences college student choices of majors and research projects. In other words, increased public literacy may help guide students into fields where their energies are most needed. Moreover, public opinion can be guided by an increased awareness of the unsolved problems in microbiology and thus influence leaders to dedicate resources to areas of need. Hence, training programs that are designed to address the grand problems of microbiology should include outreach programs that foster public and governmental awareness.
What is the best way to educate the public about micro-biology? Mechanisms for informing the public about successes in microbiology and about pressing public health issues are sorely needed.
COMMUNICATING THE ISSUES
In conveying information about microbiology to the public, it is critical first to define the target audiences and the type of information that is appropriate to convey to each audience. Potential target audiences for microbio-logical outreach comprise a long list, including business leaders, students and teachers at all levels, public officials, health professionals (who may not be sufficiently familiar with microbiology), farmers, restaurant personnel, decision-makers at federal agencies, and others.
In order to achieve the most effective outreach programs, the process of educating the public should have well-defined goals. Specific outreach programs should be conceived with specific educational goals in mind that can be implemented over a designated time period. This approach would allow targeted assessments to determine whether outreach programs were effective in communicating microbiology to the public. For example, it could be the goal of one program to educate the public on a particular microbiology topic within five to ten years, and surveys or other metrics could be used to measure the level of knowledge of the target audience.
- The intimate connections between microbial ecology and evolution, infectious disease, and the failure of standard antimicrobial therapies.
- Microbial diversity as one of the last uncharted frontiers with tremendous potential for fundamental new discoveries.
- Microbes as the foundation of the biosphere.
- The concept that the human body is nine parts microbe and one part human—for every nine microbial cells there is just one human cell.
- The tree of life and the relative placements of plants, animals, and microbes.
- The development and use of microbes as factories.
AVENUES OF COMMUNICATION
What are the best ways to convey science information to non-scientists? A number of avenues are open for outreach. For example, it may be possible to launch a campaign to present science information to the people who use public transportation: buses, trains, taxis, or in airports. The publication of popular books based on microbiological themes would also reach a significance audience. Science museums are a powerful outlet for educating young people, and interactive microbial exhibits could stimulate the minds of many future scientists in an engaging way. The mass media may also be employed. Some of these programs are already in place. For example, radio programs, like the American Society for Microbiology's “Microbe World” are well received and are proving to be highly effective.
It may be instructive to study in a systematic manner the quality of material related to microbiology that is currently being used for communication to the public. It is possible that the current lack of public savvy is due to the poor quality of information available, rather than to low availability.
Improving delivery of knowledge to the public requires engaging and informing communication professionals. Microbiological organizations should place a priority on reaching out to communication professionals and should aid in training of science writers.
Communicating with Decision-Makers
With respect to advancing the goals outlined in this report, the term “decision-makers” includes federal agencies, such as the National Science Foundation, National Institutes of Health, Environmental Protection Agency, Department of Energy, the Centers for Disease Control and Prevention, and the Department of Agriculture. Others include local and state boards of education, the Department of Homeland Security, the Food and Drug Administration, private foundations, and others.
Informed individuals who are affected by advancements in microbial science, but are not microbiologists themselves, may be among the best advocates for microbiology. Examples include representatives of biotechnology companies and their clients, business leaders who rely on the skills of highly-trained microbiologists, members of communities where property was remediated using microbes, and the beneficiaries of microbially-based therapies, including bacterially-derived antibiotics and other drugs.
The communications goals outlined in this report would be pursued most effectively by a consortium of professional societies, possibly including the American Society for Microbiology, the Society for Industrial Microbiology, the Infectious Diseases Society of America, and others.
Recommendations
Failure to acknowledge and weigh the pervasive effects of the microbial world deprives us of a powerful tool to assess the functioning of our planet and make decisions on its future as a live whole. In light of what is now known about the contributions of microorganisms to sustaining life and creating the physical and chemical properties of this planet, detailed studies in any branch of biology or geology must fully recognize the activities of microbes.
Since microbes are of fundamental importance to life and their activities must be taken into account in biology research, all biologists must have a firm background in microbial science. Coursework in micro-biology should be integrated into the core curriculum for all students in the life and earth sciences.
Building an understanding of microbes in young students will ultimately improve public awareness of the importance of microbes to the everyday health of the individual and of the planet. School science curricula in the elementary, middle, and high school levels must be amended to include lessons and lab exercises in microbiology.
The public is profoundly impacted by microbes and microbiology through disease-related matters, biotechnology, bioterrorism, and food safety. In order to improve the ability of individuals to manage their health and make informed judgments with respect to microbial science, microbiology-related professional societies should support programs that foster public microbial literacy.
- Suggested reading
- Beja, O., L. Aravind, E. V. Koonin, M. T. Suzuki, A. Hadd, L. P. Nguyen, S. B. Jovanovich, C. M. Gates, R. A. Feldman, J. L. Spudich, E. N. Spudich, and E. F. DeLong. 2000. Bacterial rhodopsin: evidence for a new type of phototrophy in the sea. Science 289:1902–1906. http://www .ncbi.nlm.nih .gov/entrez/query .fcgi?cmd=Retrieve&db =PubMed&list_uids =10988064&dopt =Abstract . [ PubMed : 10988064 ]
- Boucher, Y., C. J. Douady, R. T. Papke, D. A. Walsh, M. E. Boudreau, C. L. Nesbo, R. J. Case, and W. F. Doolittle. 2003. Lateral gene transfer and the origins of prokaryotic groups. Annual Review of Genetics 37:283–328. http://www .ncbi.nlm.nih .gov/entrez/query .fcgi?cmd=Retrieve&db =PubMed&list_uids =14616063&dopt =Abstract . [ PubMed : 14616063 ]
- Breitbart, M., I. Hewson, B. Felts, J. M. Mahaffy, J. Nulton, P. Salamon, and F. Rohwer. 2003. Metagenomic analyses of an uncultured viral community from human feces. Journal of Bacteriology 185:6220–6223. http://www .ncbi.nlm.nih .gov/entrez/query .fcgi?cmd=Retrieve&db =PubMed&list_uids =14526037&dopt =Abstract . [ PMC free article : PMC225035 ] [ PubMed : 14526037 ]
- Cohan, F. M. 2002. What are bacterial species? Annual Review of Microbiology 56:457–487. http://www .ncbi.nlm.nih .gov:80/entrez/query .fcgi?cmd=Retrieve&db =PubMed&list_uids =12142474&dopt =Abstract . [ PubMed : 12142474 ]
- Daubin, V., N. A. Moran, and H. Ochman. 2003. Phylogenetics and the cohesion of bacterial genomes. Science 301:829–832. http://www .ncbi.nlm.nih .gov/entrez/query .fcgi?cmd=Retrieve&db =PubMed&list_uids =12907801&dopt =Abstract . [ PubMed : 12907801 ]
- Delong, E. F. 2002. Microbial population genomics and ecology. Current Opinions in Microbiology 5:520–524. http://www .ncbi.nlm.nih .gov/entrez/query .fcgi?cmd=Retrieve&db =PubMed&list_uids =12354561&dopt =Abstract . [ PubMed : 12354561 ]
- Delong, E. F. 2002. Towards microbial systems science: integrating microbial perspectives, from genomes to biomes. Journal of Environmental Microbiology 4:9–10. [No abstract available.] [ PubMed : 11966814 ]
- Feil, E. J., and B. G. Spratt. 2001. Recombination and the population structures of bacterial pathogens. Annual Review of Microbiology 55:561–590. http://www .ncbi.nlm.nih .gov/entrez/query .fcgi?cmd=Retrieve&db =PubMed&list_uids =11544367&dopt =Abstract . [ PubMed : 11544367 ]
- Lerat, E., V. Daubin, and N. A. Moran. 2003. From gene trees to organismal phylogeny in prokaryotes. Proteobacteria. PLoS Biology 1:101–109. http://www .ncbi.nlm.nih .gov:80/entrez/query .fcgi?cmd=Retrieve&db =PubMed&list_uids =12975657&dopt =Abstract . Full text at http://biology .plosjournals .org/plosonline /?request=get-document&doi=10 .1371/journal .pbio.0000019 . [ PMC free article : PMC193605 ] [ PubMed : 12975657 ]
- Mann, N. H., A. Cook, A. Millard, S. Bailey, and M. Clokie. 2003. Marine ecosystems: bacterial photosynthesis genes in a virus. Nature 424:741. http://www.nature.com/cgi-taf/DynaPage.taf?file=/nature/journal/v424/n6950/abs/424741a_fs.html&dynoptions= doi1074264256 . [ PubMed : 12917674 ]
- Nelson, K. E. 2003. The future of microbial genomics. Journal of Environmental Microbiology 5:1223–1225. [No abstract available.] [ PubMed : 14641569 ]
- Ochman, H., J. G. Lawrence, and E. A. Groisman. 2000. Lateral gene transfer and the nature of bacterial innovation. Nature 405:299–304. http://www .ncbi.nlm.nih .gov/entrez/query .fcgi?cmd=Retrieve&db =PubMed&list_uids =10830951&dopt =Abstract . [ PubMed : 10830951 ]
- Rocap, G., F. W. Larimer, J. Lamerdin, S. Malfatti, P. Chain, N. A. Ahlgren, A. Arellano, M. Coleman, L. Hauser, W. R. Hess, Z. I. Johnson, M. Land, D. Lindell, A. F. Post, W. Regala, M. Shah, S. L. Shaw, C. Steglich, M. B. Sullivan, C. S. Ting, A. Tolonen, E. A. Webb, E. R. Zinser, and S. W. Chisholm. 2003. Genome divergence in two Prochlorococcus ecotypes reflects oceanic niche differentiation. Nature 424:1042–1047. http://www .ncbi.nlm.nih .gov/entrez/query .fcgi?cmd=Retrieve&db =PubMed&list_uids =12917642&dopt =Abstract . [ PubMed : 12917642 ]
- Rodriguez-Valera, F. 2002. Approaches to prokaryotic biodiversity: a population genetics perspective. Environmental Microbiology 4:628–633. http://www .ncbi.nlm.nih .gov/entrez/query .fcgi?cmd=Retrieve&db =PubMed&list_uids =12460270&dopt =Abstract . [ PubMed : 12460270 ]
- Rondon, M. R., P. R. August, A. D. Bettermann, S. F. Brady, T. H. Grossman, M. R. Liles, K. A. Loiacono, B. A. Lynch, I. A. MacNeil, C. Minor, C. L. Tiong, M. Gilman, M. S. Osburne, J. Clardy, J. Handelsman, and R. M. Goodman. 2000. Cloning the soil metagenome: a strategy for accessing the genetic and functional diversity of uncultured microorganisms. Applied and Environmental Microbiology 66:2541–2547. http://www .ncbi.nlm.nih .gov:80/entrez/query .fcgi?cmd=Retrieve&db =PubMed&list_uids =10831436&dopt =Abstract . Full text at http://aem .asm.org/cgi /content/full/66/6 /2541?view=full&pmid=10831436 . [ PMC free article : PMC110579 ] [ PubMed : 10831436 ]
- Sullivan, M. B., J. B. Waterbury, and S. W. Chisholm. 2003. Cyanophages infecting the oceanic cyanobacterium Prochlorococcus . Nature 424:1047–1051. http://www .ncbi.nlm.nih .gov/entrez/query .fcgi?cmd=Retrieve&db =PubMed&list_uids =12944965&dopt =Abstract . [ PubMed : 12944965 ]
- Tamas, I., L. Klasson, B. Canback, A. K. Naslund, A. S. Eriksson, J. J. Wernegreen, J. P. Sandstrom, N. A. Moran, and S. G. Andersson. 2002. 50 million years of genomic stasis in endosymbiotic bacteria. Science 296:2376–2379. http://www .ncbi.nlm.nih .gov/entrez/query .fcgi?cmd=Retrieve&db =PubMed&list_uids =12089438&dopt =Abstract . [ PubMed : 12089438 ]
- Ward, D. M. 1998. A natural species concept for prokaryotes. Current Opinion in Microbiology 1:271–277. http://www .ncbi.nlm.nih .gov/entrez/query .fcgi?cmd=Retrieve&db =PubMed&list_uids =10066488&dopt =Abstract . [ PubMed : 10066488 ]
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License .
- Cite this Page Microbiology in the 21st Century: Where Are We and Where Are We Going? This report is based on a colloquium sponsored by the American Academy of Microbiology held September 5–7, 2003, in Charleston, South Carolina. Washington (DC): American Society for Microbiology; 2004. doi: 10.1128/AAMCol.5Sept.2003
- PDF version of this title (227K)
In this Page
Other titles in this collection.
- American Academy of Microbiology Colloquia Reports
Related information
- NLM Catalog Related NLM Catalog Entries
Similar articles in PubMed
- Review The Global Genome Question: Microbes as the Key to Understanding Evolution and Ecology: This report is based on a colloquium, “The Global Genome Question: Microbes as the Key to Understanding Evolution and Ecology,” sponsored by the American Academy of Microbiology and held October 11-13, 2002, in Longboat Key, Florida [ 2004] Review The Global Genome Question: Microbes as the Key to Understanding Evolution and Ecology: This report is based on a colloquium, “The Global Genome Question: Microbes as the Key to Understanding Evolution and Ecology,” sponsored by the American Academy of Microbiology and held October 11-13, 2002, in Longboat Key, Florida . 2004
- Review The Uncharted Microbial World: Microbes and Their Activities in the Environment: This report is based on a colloquium, sponsored by the American Academy of Microbiology, convened February 9-11, 2007, in Seattle, Washington [ 2008] Review The Uncharted Microbial World: Microbes and Their Activities in the Environment: This report is based on a colloquium, sponsored by the American Academy of Microbiology, convened February 9-11, 2007, in Seattle, Washington . 2008
- Review Microbes in Pipes (MIP): The Microbiology of the Water Distribution System [ 2013] Review Microbes in Pipes (MIP): The Microbiology of the Water Distribution System . 2013
- Review Microbial Ecology and Genomics: A Crossroads of Opportunity: This report is based on a colloquium sponsored by the American Academy of Microbiology held February 23-25, 2001, in Singer Island, Florida [ 2002] Review Microbial Ecology and Genomics: A Crossroads of Opportunity: This report is based on a colloquium sponsored by the American Academy of Microbiology held February 23-25, 2001, in Singer Island, Florida . 2002
- Review Large-Scale Sequencing: The Future of Genomic Sciences? This report is based on a colloquium, sponsored by the American Academy of Microbiology, convened September 2008 in Washington, DC [ 2009] Review Large-Scale Sequencing: The Future of Genomic Sciences? This report is based on a colloquium, sponsored by the American Academy of Microbiology, convened September 2008 in Washington, DC . 2009
Recent Activity
- Microbiology in the 21st Century: Where Are We and Where Are We Going? Microbiology in the 21st Century: Where Are We and Where Are We Going?
Your browsing activity is empty.
Activity recording is turned off.
Turn recording back on
Connect with NLM
National Library of Medicine 8600 Rockville Pike Bethesda, MD 20894
Web Policies FOIA HHS Vulnerability Disclosure
Help Accessibility Careers
You are using an outdated browser. Please upgrade your browser or activate Google Chrome Frame to improve your experience.

What is microbiology?
Microorganisms and their activities are vitally important to virtually all processes on Earth. Microorganisms matter because they affect every aspect of our lives – they are in us, on us and around us.
Microbiology is the study of all living organisms that are too small to be visible with the naked eye. This includes bacteria, archaea, viruses, fungi, prions, protozoa and algae, collectively known as 'microbes'. These microbes play key roles in nutrient cycling, biodegradation/biodeterioration, climate change, food spoilage, the cause and control of disease, and biotechnology. Thanks to their versatility, microbes can be put to work in many ways: making life-saving drugs, the manufacture of biofuels, cleaning up pollution, and producing/processing food and drink.
Microbiologists study microbes, and some of the most important discoveries that have underpinned modern society have resulted from the research of famous microbiologists, such as Jenner and his vaccine against smallpox, Fleming and the discovery of penicillin, Marshall and the identification of the link between Helicobacter pylori infection and stomach ulcers, and zur Hausen, who identified the link between papilloma virus and cervical cancer.
Microbiology research has been, and continues to be, central to meeting many of the current global aspirations and challenges, such as maintaining food, water and energy security for a healthy population on a habitable earth. Microbiology research will also help to answer big questions such as 'how diverse is life on Earth?', and 'does life exist elsewhere in the Universe'?
Introducing microbes
More than just pathogens - can be friend or foe.
Smallest of all the microbes, but are they alive?
More than just mushrooms.
Microbes with a taste for poo and so much more.
Microbial powerhouses essential for life.
First found existing on the edge of life.
Mysterious misfolding proteins.
Microbes in the world
Microbes and the human body.
Ever wondered why when we are surrounded by microbes we are not ill all the time?
Microbes and food
Food for thought – bread, chocolate, yoghurt, blue cheese and tofu are all made using microbes.
Microbes and the outdoors
The function of microbes as tiny chemical processors is to keep the life cycles of the planet turning.
Microbes and climate change
How are microbes contributing to climate change?

General Microbiology - 1st Edition
(6 reviews)
Linda Bruslind, Oregon State University
Copyright Year: 2020
Publisher: Oregon State University
Language: English
Formats Available
Conditions of use.
Learn more about reviews.
Reviewed by Rajeev Chandra, Assistant Professor, Norfolk State University on 5/9/23
This first edition of Professor Bruslind's well-established textbook is an excellent introduction to microbiology to a wide range of undergraduate students. Although primarily aimed at students of microbiology, it will also be suitable for all... read more
Comprehensiveness rating: 4 see less
This first edition of Professor Bruslind's well-established textbook is an excellent introduction to microbiology to a wide range of undergraduate students. Although primarily aimed at students of microbiology, it will also be suitable for all life-science undergraduates whose courses include microbiology as a component, whether they are studying botany, zoology, pharmacy, medicine or agriculture as their main subject. By concentrating on the essential aspects of microbiology, the student is unerringly provided with an excellent general overview of microbiology, without becoming distracted by a wealth of unnecessary detail. This new edition brings fully up to date the various advances in the subject, including new species, phylogenetic relationships and novel metabolic pathways, and a greater emphasis on environmental and ecological matters.
Content Accuracy rating: 5
• The engaging tools in this book assist students in practicing, visualizing and comprehend important content. • Focuses on learners by the inclusion of research-based activities, engaging activities, case studies to improve the ability of students to solve problems and connect between concepts. • Teaches tough topics in microbiology with superior art and illustrations. The micrographs enable students to digest difficult microbiology processes and concepts.
Relevance/Longevity rating: 4
• It proceeds in setting up the standard for impeccable scholarship and accuracy, as well as outstanding photos and illustrations. • This textbook is meant for microbiology, biology, and several other science majors. • It balances cutting-edge research with concepts that are essential for understanding the microbiology field, including strong coverage of evolution, ecology, and metabolism. However the text is missing in some exhaustive Epidemiological data, if added can be excellent text.
Clarity rating: 5
• The textbook offers a rock-solid basic foundation in microbiology, and it clearly and carefully explains the fundamental concepts through an offering of appealing and vivid instructional art. • The book always draws back students to it again and again due to the nature of the text and excellent explanations. • The text has a readable and concise style, covers current concepts, and provides students the mastery and knowledge necessary to comprehend future advances. • The coverage of diseases uses a body systems approach.
Consistency rating: 5
• The information and accompanying resources help students in making connections between the theory of microbiology and disease diagnosis, prevention, and treatment. • Regardless of whether your study caters to microbiology majors, pre-health professional students, or pre-med students, everything that is needed for a thorough introduction of microbiology is available in this book.
Modularity rating: 5
The text has a readable and concise style, covers current concepts, and provides students the mastery and knowledge necessary to comprehend future advances.
Organization/Structure/Flow rating: 5
• It proceeds in setting up the standard for impeccable scholarship and accuracy, as well as outstanding photos and illustrations. • This textbook is meant for microbiology, biology, and several other science majors. • It balances cutting-edge research with concepts that are essential for understanding the microbiology field, including strong coverage of evolution, ecology, and metabolism.
Interface rating: 5
The text is free of significant interface issues, including navigation problems, distortion of images/charts, and any other display features that may distract or confuse the reader.
Grammatical Errors rating: 5
The text contains no grammatical errors.
Cultural Relevance rating: 5
The text is not culturally insensitive or offensive in any way. It should make use of examples that are inclusive of a variety of races, ethnicities, and backgrounds.
Reviewed by Katherine Buhrer, Assistant Professor, Biology, Tidewater Community College on 4/25/23
The areas that were covered in the text were well organized and addressed in a succinct manner. I would like to see microbes other than bacteria covered in order to make the book more useful in a general introductory microbiology course. The... read more
The areas that were covered in the text were well organized and addressed in a succinct manner. I would like to see microbes other than bacteria covered in order to make the book more useful in a general introductory microbiology course. The book is limited to bacteria and viruses in general. would also like to see the immunological response to microbes addressed, as well as some diseases of the systems if I were going to use the text in the Microbiology course I teach. I did not see a glossary or index, both of which are usually quite helpful to students.
Content Accuracy rating: 4
I found the book to be accurate in the material that was presented, I did not note any errors in content. Diagrams were generally large enough to see detail and well labeled. One exception, the diagram of BLAST results was difficult to read, perhaps because I downloaded it as a PDF, maybe the electronic version would be clearer.
Relevance/Longevity rating: 5
The book has a lot of general information on microbes that is good basic information; current and basic enough to not likely be outdated - more information may easily be added in the future but what is in the text will not likely change.
The book is written almost like a conversation, I think students will find it interesting to read as a result of this. Terminology is relevant, lending a good vocabulary base to the reader.
The book is very consistent in depth of content, presentation and organization of material. Each chapter is well organized and follows up with a terminology "box" as well as questions to encourage students to focus on the key components of each section.
One of the things I really liked about this book is the length of sections of material. The author did a great job of presenting topics in very concise, readable sections. I think this keeps the reader focused better than long, monotonous paragraphs that are loaded with more detail than is needed.
Organization/Structure/Flow rating: 4
The order of topics was satisfactory, the only exception was viruses were in two separate sections, I would probably have combined the sections.
Interface rating: 4
The only exception to being absolutely fine in this category was the one diagram mentioned earlier, the BLAST results diagram - it came out as a little blurry in the PDF version of the book.
I did not note any grammatical errors.
I did not note any material that was insensitive or offensive in any way.
I thought the book was extremely thorough in the sections on prokaryotic cell structure. It provides more than enough material for the instructor to cover the topic to the degree they wish. The microbe groups the book covers, are covered very well (bacteria and viruses) but I think to be useful as an Introductory Microbiology text, I would like to see some of the other microbe groups included.
Reviewed by Andrea Beyer, Assistant Professor, Virginia State University on 8/22/21
This text covers a broad range of the introductory microbiology basics in a clean, straight-to-the point manner, and introduces the working vocabulary needed. It covers many core topics that I would expect to find in a micro class, with the... read more
This text covers a broad range of the introductory microbiology basics in a clean, straight-to-the point manner, and introduces the working vocabulary needed. It covers many core topics that I would expect to find in a micro class, with the exception of material discussing control of microbial growth (disinfectants/autoclaving/antibiotics), antibiotic resistance (how it is acquired, why it is a problem), and host-pathogen interactions (related to human health/immunity/vaccinations/medicine). A little more could be included about eukaryotic microbes, but most micro classes do focus more on prokaryotes, which this covers. Most of the American Society for Microbiology Core Competencies for Undergraduate Microbiology are met with the few exceptions as noted above.
The content that I reviewed was accurate and expected for an intro to micro text. For the very few lesser-known pieces of information that were conversationally shared, a reference or link to cite the source would be helpful for readers who are curious! (one example- page 76: “All good things must come to an end (otherwise bacteria would equal the mass of the Earth in 7 days!)”- how was that was calculated?)
As written, it largely contains the foundations of microbiology, and a small bit of historical information, that is unlikely to change within the next few years. The exception would be the end chapters covering genetic engineering (as new techniques evolve) and microbial genomics (as DNA sequencing expands our current view of microbial genomes).
Clarity rating: 4
The writing style is more conversational, which can take a little getting used to, compared to many other textbook stylings that sound more formal and scientific. This means the tone can be a little casual, but I don’t think it is distracting; it makes for a very approachable view of Microbiology that is appealing to students, especially non-major undergrads.
The text flows in the same conversational pattern throughout, giving one voice to the flow of information. Each chapter has the same formatting, with the content followed by a box of keywords (which is useful as a summary of key chapter terms/names/topics for students) and study questions directly related to what was just read. All vocabulary words are noted in bold throughout the text.
I enjoyed the overall setup of this text; the sections cover the essentials without extra fluff- and the sections are not long and cumbersome. They cover the basics, and move on. This could serve as a backbone structure for a class with other supplemental materials provided to accompany it (such as case studies, articles, videos, etc).
Going from the start and moving towards the back of the book, the chapters progress in their content and build upon the previous information. This is the kind of book that almost anyone could pick up and guide themself through a working knowledge of microbes.
I easily read this as a PDF file and on the website. Most pictures are large and well-colored, and nicely demonstrate the ideas discussed. A few images here and there are pixelated/a little blurry, but not to the point of not being able to see the object/main idea. All links that I tested worked and went to relevant/appropriate websites.
Rare- did not encounter many (1-2 per chapter at most).
Cultural Relevance rating: 4
The text is very straight forward, and I did not see any instances of insensitive examples or statements as it is centralized to the microorganisms. (see overall comment)
This book focuses primarily on the microbial aspect of microbiology, without major emphasis on the human perspective of it (health/historically/culturally). While this makes the text very simple to read and understand, it is missing that connection of the microbial world to the human one. And this might make it harder for students to see how important and impactful microbes are in our world. Examples of microbiology impacting daily life could give relevance and context to much of the material presented in this text.
Reviewed by Roger Greenwell, Associate Professor, Worcester State University on 6/30/21
The focus of this concise and easy to read text is the introduction of bacteria, archaea, and viruses and how they function and are controlled. Fungi and other microbes are minimally included. The effects of microbes on humans, the function of... read more
Comprehensiveness rating: 3 see less
The focus of this concise and easy to read text is the introduction of bacteria, archaea, and viruses and how they function and are controlled. Fungi and other microbes are minimally included. The effects of microbes on humans, the function of the immune system, and disease principles and vaccination are not present. For the content presented in the 22 chapters, it would be easy for an instructor to supplement additional content to enhance the course content with literature, case studies, and more. The later chapters that have some of the content that is subject to more frequent change/updating (such as microbial engineering) but there is some opportunity to add some additional info (such as on DNA sequencing, it would be nice to see Sanger Sequencing and a figure on that included, and a figure on how CRISPR-Cas9 works in bacteria). All of the terminology that is necessary is bolded and included at the end of each chapter. Additional links to supplemental information is useful to expand the information. The questions at the end of each chapter highlight the expected learning objectives from each chapter.
Content Accuracy rating: 3
Much of the content is accurate through the text, but some of the explanations are simplified to the point of inaccuracy (for example, the description of peptidoglycan indicates that L-amino acids are primarily used by cells -- it should be clarified that they are primarily used by cells for protein synthesis, as there are several cellular uses for D-amino acids in bacteria including peptidoglycan, secondary metabolite production, and quorum sensing). There are some term associations that are not commonly used (for example, classifying spirochaetes as curved rods); incorrect terms used (the singular for pili is pilus, not pilin); some missing information that should be included (the bacterial morphology should include diplococci, streptococci, hyphal structures, and others). Some of the figures need some correction: the phylogenetic trees are different and indicate that fungi are more closely related to plants (fungi and animals are more closely related, part of the Opisthokonta supergroup); the prokaryotic cell has the fimbriae labeled as pili.
Relevance/Longevity rating: 3
This text is written to give the basics of microbiology throughout. While the field of microbiology is ever-changing, this book is mostly sufficient to give the foundations to students, and instructors can easily supplement with more specific information, case studies, etc. However, there is no content about the immune system, which should be included for majors microbiology or medical microbiology courses.
The text is written for ease of reading; the conversational tone helps with accessibility to the information presented by the authors. Some of the more complicated explanations have figures to help with visualizing the key points, and the embedded links assist in further clarifying the content.
Consistency rating: 4
Each chapter has a consistent, conversational tone that should allow students to connect easily with the content. Important terms are bolded and defined throughout the entire text, and then listed at the end of each chapter as key words. The figures are sufficiently easy to follow, though I would recommend that the figures have standalone descriptions. One inconsistency in early chapters is the introduction of some terms with the initial plural term (singular in parentheses), whereas later chapters do the opposite.
This text is easily modular and can be rearranged into various subunits based on the course structure as needed by the instructor. While designed in an overall logical order, one could easily move content to focus on different topics at various points in the semester.
The overall organization is mostly logical and straightforward. I would recommend moving the content in Chapter 16 earlier in the text because it introduces Early Earth and evolution, and defines phylogenetic trees; however multiple phylogenetic trees are introduced in earlier chapters.
Most of the figures were easily identifiable and clear. In the PDF version of the text, the cell wall structures image in Chapter 7 is blurry and needs to be fixed. The embedded links were functional and provided nice additional resources to the students as they go through the content.
Grammatical Errors rating: 4
Overall the grammar is consistent and there are limited errors that stand to be corrected.
The focus of this text is primarily on the microbes themselves and how they grow. There is no focus on culture or ethnicity. There is a missed opportunity to connect microbiology to their greater impact on the planet and humanity, such as the impacts of microbes on human evolution and human history. The history of microbiology should be included to provide some additional cultural context.
Overall this is an easy-to-read, basic microbiology textbook for students. If the content corresponds to all of the course SLOs, then this would be a good text to use.
Reviewed by Sarah Olken, Professor, North Shore Community College on 6/29/21
As noted in the first chapter, Eukaryotic microorganisms are not covered in detail. Host Immunity and Vaccination are not covered. These omissions are a weakness of the text, in my opinion. read more
As noted in the first chapter, Eukaryotic microorganisms are not covered in detail. Host Immunity and Vaccination are not covered. These omissions are a weakness of the text, in my opinion.
Content is accurate.
Chapters covered topics relevant to Microbiology.
Very concise and clear. Bruslind’s writing style is very accessible which is a strength of the text. I expect students will appreciate the concise language that sticks to main points. I anticipate students will rely on prior knowledge for some chapters. For example, there is no review of the chemistry of life and biomolecules. Other chapters seem incomplete. For example, Chapter 21 defines virulence as the measure of a pathogen’s likelihood to cause disease but does not explain how virulence is measured. This may mean students need to use other sources to fully understand content.
Great format. Each chapter format includes section titles, a list of key terms and study questions that align with common learning objectives.
Modularity rating: 4
Chapters are nicely parsed, with the exception of Chapter 21. In my opinion this chapter should be split into 2 chapters, one covering epidemiology and the other covering pathogenicity in more detail (more about virulence factors) than provided in this chapter. I appreciate the separation of Microbial Metabolism into several chapters.
Great flow that is similar to most Microbiology texts. The separation of bacterial cell wall and external structures into two chapters is a great way to avoid confusion about the cell envelope. I appreciate the thorough introduction to Bacteria, Archaea and viruses before covering microbial growth and metabolism.
No problem as viewed on Google Chrome.
No errors found. Writing style is clear.
Cultural Relevance rating: 3
This is a weakness of the text. History of Microbiology is Eurocentric. Example of nomenclature uses a European male name. CRIPSR-Cas is explained without mention of Emmanuelle Charpentier or Jennifer Doudna, both of whom won the Nobel prize for its discovery.
Note that as the title suggests, this is a general text, providing the ‘bones’ of Microbiology. I see it as more appropriate for non-majors course at 100 level. The style of writing is very approachable and even informal at times. The strength is the concise way topics are presented. This is also the limitation of the text for a few reasons. First, there are less figures and/or simplified figures which is a weakness, especially for visual learners. Second, there is little connection to practical examples. However, this can be supplemented by classwork. Third, there are no chapters covering human infectious diseases (as are usually included in Microbiology textbooks for Allied Health). Since information about human infectious diseases changes frequently, this may be seen as an advantage, giving the instructor the ability to update the course with current topics. In addition, this means that the text may be used for a course in Environmental Microbiology, Food Microbiology (with the addition of material on yeast), or other special topic in Microbiology. I would find the text useful for those looking for a basic text upon which they can customize examples and explore the relevance of each topic through classroom activities and facilitated discussions.
Reviewed by Jacqueline Spencer, Assistant Professor of Biology, Thomas Nelson Community College on 5/30/21
This is a very concisely written, comprehensive textbook that covers most of the major topics usually found in a General Microbiology course. The key word here is “general” because this is not a text that is specifically written for Health... read more
Comprehensiveness rating: 5 see less
This is a very concisely written, comprehensive textbook that covers most of the major topics usually found in a General Microbiology course. The key word here is “general” because this is not a text that is specifically written for Health Sciences students who will be going into nursing, dental hygiene, etc., although there is one very short chapter on Bacterial Pathogenicity and another on viral transmission. However, there is little mention of other fungal or helminth infections. The interactive “Contents” that appears on the left of the screen when needed, lists the contents of the 22 individual chapters. Each chapter title serves as a “hot link” that takes the reader to that page within the textbook. Helpful features for both instructors and students are the “Key Words” and “Study Questions” that are posted at the conclusion of every chapter. Additionally, the key words are set in eye-catching bold type throughout the text. The chapters are surprisingly short in length given what is usually seen in the average lengthy textbook for this complex course. But the author has chosen to convey the critical information that a student needs without all of the wordiness that is more common in other texts. Within each chapter, the topic is covered in a conversational way that engages the reader because of the friendly, modern and often humorous way it is written. It reads like what an engaging lecturer would be saying in an actual classroom. There are also pertinent figures and illustrations to help understanding of critical concepts.
The thrifty use of words in the chapters that demand highly technical explanations and/or descriptions doesn’t lend itself to many inaccuracies because detailed information is kept “short and sweet”. So, there are no glaring errors that could be detected. Instead, the details are very straightforward and presented in a readable and conversational language. In the one chapter on Genomics that contains very detailed technological explanations, there were no errors detected.
Because Microbiology is a field of biology that is always undergoing change (like the Covid-19 Pandemic!), many textbooks are invariably out-of-date from the aspect of new technologies and microbial strains included from the moment they are published. The abbreviated format of this text skirts this issue by providing minimal but accurate information on more recent technologies and these are found in the chapter on Genomics, where only one or two paragraphs is used to describe them along with helpful diagrams and figures where needed. Instructors using this text would find it easy to supplement more recent material as necessary
This text is written in a conversational type of prose which normally might not lend itself to scientific technical jargon, but that is not of concern here because topics are described concisely. Complicated information is explained clearly for ease in understanding.
This 1st Edition of this text is very consistent as to its framework and level of terminology used. Each of the 22 chapters is concise, offering basic diagrams and/or figures as necessary. Important terminology is printed in bold typeface and included in a list of key words at the chapter’s end along with the study questions. There is an evenness to the entire text.
This text exhibits its modularity in the chapters themselves, rather than in parts or sections, because the chapters are so concise. These chapters can easily be arranged to complement the organization of a particular course. With 22 different chapters on specific aspects of microbial life available in compact units of information, many different sequences can be chosen by an instructor. An example of the concentration of information in the individual chapters is the one on Genomics that is like a miniature encyclopedia of biotechnology involving microbes.
This concise text on what can be the overpowering topic of General Microbiology demonstrates good organization in the ordering of the chapter topics. Because this is a “General” Microbiology text, the main themes are the differences between the different types of microbes according to their genomes, their metabolism and their relationships to the rest of our global ecology.
There are no apparent issues with the interface between sections of this text. All figures and diagrams were clearly visible and it was very easy to navigate between individual chapters as well as find them in the content section.
There did not appear to be any issues with either grammar or spelling in any of the chapters.
Because the entire text is about microorganisms and there is little or no reference to humans, there are no problems with any cultural aspects. There is nothing in the subject matter that could be construed as being either insensitive or offensive to any students using it.
This is a very good General Microbiology text that covers all the bases. Additionally, students should like its conversational tone as the author “speaks” to the reader with some humor and wit when warranted, thus drawing the reader in as a participant in the discussion rather than as an observer. It is based on sound scientific facts about the wide range of microorganisms on our planet. The only thing it lacks is the history of Microbiology and any real mention of the scientists who contributed to our current vast knowledge.
Table of Contents
- 1. Introduction to Microbiology
- 2. Microscopes
- 3. Cell Structure
- 4. Bacteria: Cell Walls
- 5. Bacteria: Internal Components
- 6. Bacteria: Surface Structures
- 8. Introduction to Viruses
- 9. Microbial Growth
- 10. Environmental Factors
- 11. Microbial Nutrition
- 12. Energetics & Redox Reactions
- 13. Chemoorganotrophy
- 14. Chemolithotrophy & Nitrogen Metabolism
- 15. Phototrophy
- 16. Taxonomy & Evolution
- 17. Microbial Genetics
- 18. Genetic Engineering
- 19. Genomics
- 20. Microbial Symbioses
- 21. Bacterial Pathogenicity
- 22. The Viruses
Ancillary Material
About the book.
Welcome to the wonderful world of microbiology! Yay! So. What is microbiology? If we break the word down it translates to “the study of small life,” where the small life refers to microorganisms or microbes. But who are the microbes? And how small are they? Generally microbes can be divided in to two categories: the cellular microbes (or organisms) and the acellular microbes (or agents). In the cellular camp we have the bacteria, the archaea, the fungi, and the protists (a bit of a grab bag composed of algae, protozoa, slime molds, and water molds). Cellular microbes can be either unicellular, where one cell is the entire organism, or multicellular, where hundreds, thousands or even billions of cells can make up the entire organism. In the acellular camp we have the viruses and other infectious agents, such as prions and viroids. In this textbook the focus will be on the bacteria and archaea (traditionally known as the “prokaryotes,”) and the viruses and other acellular agents.
About the Contributors
Linda Bruslind , Oregon State University
Contribute to this Page
Thank you for visiting nature.com. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser (or turn off compatibility mode in Internet Explorer). In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript.
- View all journals
- Explore content
- About the journal
- Publish with us
- Sign up for alerts
- Published: 08 April 2024
A dual-targeting antifungal is effective against multidrug-resistant human fungal pathogens
- Min Zhou ORCID: orcid.org/0000-0002-0444-0324 1 , 2 na1 ,
- Longqiang Liu 2 na1 ,
- Zihao Cong ORCID: orcid.org/0000-0003-3462-7373 2 ,
- Weinan Jiang 2 ,
- Ximian Xiao ORCID: orcid.org/0000-0002-7096-8784 2 ,
- Jiayang Xie 2 ,
- Zhengjie Luo 2 ,
- Sheng Chen ORCID: orcid.org/0000-0001-9090-3905 2 ,
- Yueming Wu 2 ,
- Xinying Xue 3 , 4 ,
- Ning Shao 2 &
- Runhui Liu ORCID: orcid.org/0000-0002-7699-086X 1 , 2
Nature Microbiology ( 2024 ) Cite this article
842 Accesses
18 Altmetric
Metrics details
- Biomaterials
- Drug discovery and development
Drug-resistant fungal infections pose a significant threat to human health. Dual-targeting compounds, which have multiple targets on a single pathogen, offer an effective approach to combat drug-resistant pathogens, although ensuring potent activity and high selectivity remains a challenge. Here we propose a dual-targeting strategy for designing antifungal compounds. We incorporate DNA-binding naphthalene groups as the hydrophobic moieties into the host defence peptide-mimicking poly(2-oxazoline)s. This resulted in a compound, (Gly 0.8 Nap 0.2 ) 20 , which targets both the fungal membrane and DNA. This compound kills clinical strains of multidrug-resistant fungi including Candida spp., Cryptococcus neoformans , Cryptococcus gattii and Aspergillus fumigatus . (Gly 0.8 Nap 0.2 ) 20 shows superior performance compared with amphotericin B by showing not only potent antifungal activities but also high antifungal selectivity. The compound also does not induce antimicrobial resistance. Moreover, (Gly 0.8 Nap 0.2 ) 20 exhibits promising in vivo therapeutic activities against drug-resistant Candida albicans in mouse models of skin abrasion, corneal infection and systemic infection. This study shows that dual-targeting antifungal compounds may be effective in combating drug-resistant fungal pathogens and mitigating fungal resistance.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
24,99 € / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
111,21 € per year
only 9,27 € per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
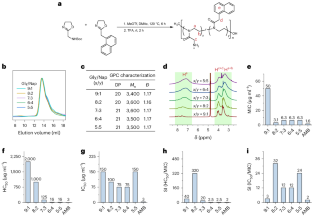
Similar content being viewed by others
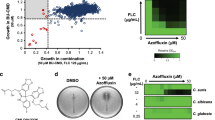
An oxindole efflux inhibitor potentiates azoles and impairs virulence in the fungal pathogen Candida auris
Kali R. Iyer, Kaddy Camara, … Leah E. Cowen
Molecular mechanisms governing antifungal drug resistance
Yunjin Lee, Nicole Robbins & Leah E. Cowen

Drug resistance and tolerance in fungi
Judith Berman & Damian J. Krysan
Data availability
Data supporting the findings of this study are available within the Article and Supplementary Information . Source data are provided with this paper. Any other source data perceived as pertinent are available, on reasonable request, from the corresponding author.
Fukushima, K. et al. Supramolecular high-aspect ratio assemblies with strong antifungal activity. Nat. Commun. 4 , 2861 (2013).
Article PubMed Google Scholar
Pappas, P. G., Lionakis, M. S., Arendrup, M. C., Ostrosky-Zeichner, L. & Kullberg, B. J. Invasive candidiasis. Nat. Rev. Dis. Primers 4 , 18026 (2018).
Kupferschmidt, K. New drugs target growing threat of fatal fungi. Science 366 , 407 (2019).
Article CAS PubMed Google Scholar
Zhang, X. et al. Development of lipo-γ-AA peptides as potent antifungal agents. J. Med. Chem. 65 , 8029–8039 (2022).
Fisher, M. C., Hawkins, N. J., Sanglard, D. & Gurr, S. J. Worldwide emergence of resistance to antifungal drugs challenges human health and food security. Science 360 , 739–742 (2018).
Zhang, F. et al. A marine microbiome antifungal targets urgent-threat drug-resistant fungi. Science 370 , 974–978 (2020).
Article CAS PubMed PubMed Central Google Scholar
Fisher, M. C. et al. Tackling the emerging threat of antifungal resistance to human health. Nat. Rev. Microbiol. 20 , 557–571 (2022).
Berman, J. & Krysan, D. J. Drug resistance and tolerance in fungi. Nat. Rev. Microbiol. 18 , 319–331 (2020).
Iyer, K. R., Revie, N. M., Fu, C., Robbins, N. & Cowen, L. E. Treatment strategies for cryptococcal infection: challenges, advances and future outlook. Nat. Rev. Microbiol. 19 , 454–466 (2021).
Lockhart, S. R., Chowdhary, A. & Gold, J. A. W. The rapid emergence of antifungal-resistant human-pathogenic fungi. Nat. Rev. Microbiol. 21 , 818–832 (2023).
Day, J. N. et al. Combination antifungal therapy for cryptococcal meningitis. N. Engl. J. Med. 368 , 1291–1302 (2013).
Shekhar-Guturja, T. et al. Dual action antifungal small molecule modulates multidrug efflux and TOR signaling. Nat. Chem. Biol. 12 , 867–875 (2016).
Theuretzbacher, U. Dual-mechanism antibiotics. Nat. Microbiol. 5 , 984–985 (2020).
Silver, L. L. Multi-targeting by monotherapeutic antibacterials. Nat. Rev. Drug Discov. 6 , 41–55 (2007).
Parkes, A. L. & Yule, I. A. Hybrid antibiotics—clinical progress and novel designs. Expert Opin. Drug Discov. 11 , 665–680 (2016).
Yarlagadda, V., Sarkar, P., Samaddar, S. & Haldar, J. A vancomycin derivative with a pyrophosphate-binding group: a strategy to combat vancomycin-resistant bacteria. Angew. Chem. Int. Ed. Engl. 55 , 7836–7840 (2016).
Jiang, Y. et al. “Metaphilic” cell-penetrating polypeptide–vancomycin conjugate efficiently eradicates intracellular bacteria via a dual mechanism. ACS Cent. Sci. 6 , 2267–2276 (2020).
Martin, J. K. et al. A dual-mechanism antibiotic kills gram-negative bacteria and avoids drug resistance. Cell 181 , 1518–1532 (2020).
Bai, S. et al. A polymeric approach toward resistance-resistant antimicrobial agent with dual-selective mechanisms of action. Sci. Adv. 7 , eabc9917 (2021).
Denning, D. W. & Bromley, M. J. How to bolster the antifungal pipeline. Science 347 , 1414–1416 (2015).
Kuroda, K. & DeGrado, W. F. Amphiphilic polymethacrylate derivatives as antimicrobial agents. J. Am. Chem. Soc. 127 , 4128–4129 (2005).
Hancock, R. E. & Sahl, H. G. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat. Biotechnol. 24 , 1551–1557 (2006).
Xiong, M. et al. Helical antimicrobial polypeptides with radial amphiphilicity. Proc. Natl Acad. Sci. USA 112 , 13155–13160 (2015).
Lee, E. Y. et al. Helical antimicrobial peptides assemble into protofibril scaffolds that present ordered dsDNA to TLR9. Nat. Commun. 10 , 1012 (2019).
Article PubMed PubMed Central Google Scholar
Mookherjee, N., Anderson, M. A., Haagsman, H. P. & Davidson, D. J. Antimicrobial host defence peptides: functions and clinical potential. Nat. Rev. Drug Discov. 19 , 311–332 (2020).
Xian, W. et al. Histidine-mediated ion specific effects enable salt tolerance of a pore-forming marine antimicrobial peptide. Angew. Chem. Int. Ed. Engl. 61 , e202108501 (2022).
Sang, P. & Cai, J. Unnatural helical peptidic foldamers as protein segment mimics. Chem. Soc. Rev. 52 , 4843–4877 (2023).
Hurdle, J. G., O’Neill, A. J., Chopra, I. & Lee, R. E. Targeting bacterial membrane function: an underexploited mechanism for treating persistent infections. Nat. Rev. Microbiol. 9 , 62–75 (2011).
Nederberg, F. et al. Biodegradable nanostructures with selective lysis of microbial membranes. Nat. Chem. 3 , 409–414 (2011).
Krumm, C. et al. Antimicrobial poly(2-methyloxazoline)s with bioswitchable activity through satellite group modification. Angew. Chem. Int. Ed. Engl. 53 , 3830–3834 (2014).
Bai, H. et al. A supramolecular antibiotic switch for antibacterial regulation. Angew. Chem. Int. Ed. Engl. 54 , 13208–132013 (2015).
Lam, S. J. et al. Combating multidrug-resistant Gram-negative bacteria with structurally nanoengineered antimicrobial peptide polymers. Nat. Microbiol. 1 , 16162 (2016).
Chin, W. et al. A macromolecular approach to eradicate multidrug resistant bacterial infections while mitigating drug resistance onset. Nat. Commun. 9 , 917 (2018).
Zhong, W. et al. Designer broad-spectrum polyimidazolium antibiotics. Proc. Natl Acad. Sci. USA 117 , 31376–31385 (2020).
Cho, Y. et al. Noncovalent interactions of DNA bases with naphthalene and graphene. J. Chem. Theory Comput. 9 , 2090–2096 (2013).
Zhou, M. et al. Poly(2-oxazoline)-based functional peptide mimics: eradicating MRSA infections and persisters while alleviating antimicrobial resistance. Angew. Chem. Int. Ed. Engl. 59 , 6412–6419 (2020).
Whitfield, C. J. et al. Functional DNA–polymer conjugates. Chem. Rev. 121 , 11030–11084 (2021).
Rautenbach, M., Troskie, A. M. & Vosloo, J. A. Antifungal peptides: to be or not to be membrane active. Biochimie 130 , 132–145 (2016).
Jiang, W. et al. Short guanidinium-functionalized poly(2-oxazoline)s displaying potent therapeutic efficacy on drug-resistant fungal infections. Angew. Chem. Int. Ed. Engl. 61 , e202200778 (2022).
Dos Reis, T. F. et al. A host defense peptide mimetic, brilacidin, potentiates caspofungin antifungal activity against human pathogenic fungi. Nat. Commun. 14 , 2052 (2023).
Chang, W. et al. Efflux pump-mediated resistance to antifungal compounds can be prevented by conjugation with triphenylphosphonium cation. Nat. Commun. 9 , 5102 (2018).
Download references
Acknowledgements
This study was supported by the National Natural Science Foundation of China (number T2325010 (R.L.)), National Key Research and Development Program of China (2022YFC2303100 (R.L.)), National Natural Science Foundation of China (22075078 (R.L.), 22305082 (M.Z.), 52203162 (Y.W.), 82370005 (X.X.)), Shanghai Frontiers Science Center of Optogenetic Techniques for Cell Metabolism (Shanghai Municipal Education Commission) (R.L.), Open Research Fund of State Key Laboratory of Polymer Physics and Chemistry (Changchun Institute of Applied Chemistry, Chinese Academy of Sciences) (R.L.) and the Open Project of Engineering Research Center of Dairy Quality and Safety Control Technology (Ministry of Education, R202201) (R.L.). We thank the Analysis and Testing Center of the East China University of Science and Technology for their help in the characterization. We also thank the Analysis and Testing Center of the School of Chemical Engineering, East China University of Science and Technology for the support. We thank the staff members of the Integrated Laser Microscopy System at the National Facility for Protein Science in Shanghai (NFPS), Zhangjiang Lab, China, for providing technical support and assistance in data collection and analysis.
Author information
These authors contributed equally: Min Zhou, Longqiang Liu.
Authors and Affiliations
State Key Laboratory of Bioreactor Engineering, East China University of Science and Technology, Shanghai, China
Min Zhou & Runhui Liu
Shanghai Frontiers Science Center of Optogenetic Techniques for Cell Metabolism, Key Laboratory for Ultrafine Materials of Ministry of Education, Frontiers Science Center for Materiobiology and Dynamic Chemistry, Research Center for Biomedical Materials of Ministry of Education, School of Materials Science and Engineering, East China University of Science and Technology, Shanghai, China
Min Zhou, Longqiang Liu, Zihao Cong, Weinan Jiang, Ximian Xiao, Jiayang Xie, Zhengjie Luo, Sheng Chen, Yueming Wu, Ning Shao & Runhui Liu
Department of Respiratory and Critical Care, Beijing Shijitan Hospital, Capital Medical University, Beijing, China
Xinying Xue
School of Clinical Medicine, Shandong Second Medical University, Weifang, China
You can also search for this author in PubMed Google Scholar
Contributions
M.Z. and R.L. conceived the idea, proposed the strategy, designed the experiments, evaluated the data and wrote the paper together. M.Z. performed majority of the experiments. L.L., Z.C. and Z.L. contributed to the in vivo animal models. W.J., X.X. and S.C. participated in the confocal microscopy study. J.X. performed the SEM test. Y.W. contributed to the data analysis and result discussion. N.S. contributed to the cytotoxicity assay. X.X. contributed to the antifungal activity test on Cryptococcus gatti . All authors proofread the paper.
Corresponding author
Correspondence to Runhui Liu .
Ethics declarations
Competing interests.
R.L. and M.Z. are co-inventors of a patent application covering the reported synthesis of poly(2-oxazoline)s and their antifungal application. The other authors declare no competing interests.
Peer review
Peer review information.
Nature Microbiology thanks Dominique Sanglard, Shu Wang and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended data fig. 1 design and performance of dual-targeting antifungal hdp-mimicking poly(2-oxazoline)s..
(a) Design of dual-targeting HDP-mimicking poly(2-oxzoline)s bearing amphiphilic structure for membrane targeting and naphthalene group for DNA binding. (b) Schematic diagram of dual-targeting and dual-mechanism antifungal poly(2-oxzoline)s targeting both fungal cell membrane and DNA. (c) Potent therapeutic effect of the optimal antifungal poly(2-oxzoline) in treating mice drug-resistant fungal infections as demonstrated using multiple animal models.
Extended Data Fig. 2 Fungicidal efficiency of (Gly 0.8 Nap 0.2 ) 20 and corresponding PI staining of fungi.
(a) The percentage of killed Candida albicans K1 treated with (Gly 0.8 Nap 0.2 ) 20 at 1×MFC concentration, with an initial concentration of 1×10 6 CFU/mL in RPMI 1640 medium. n = 3 independent samples. Data are presented as mean values ± SD. (b-f) Confocal images and the corresponding percentage of PI staining of (Gly 0.8 Nap 0.2 ) 20 -treated Candida albicans K1 at variable time points, with an initial fungal concentration of 1×10 6 CFU/mL in RPMI 1640 medium. The confocal imaging was collected once, at least 50 fungal cells were observed individually in the sample displaying results similar to the representative images as shown in this figure.
Source data
Extended data fig. 3 influence of exogenous fungal membrane lipid components on the antifungal activity of (gly 0.8 nap 0.2 ) 20..
Change of (Gly 0.8 Nap 0.2 ) 20 ’s antifungal activity in the presence of exogenous fungal membrane lipid components (a) phosphatidylglycerol (PG) and (b) phosphatidylinositol (PI), respectively, using terbinafine for comparison.
Extended Data Fig. 4 Influence of intracellular ROS on the antifungal activity of (Gly 0.8 Nap 0.2 ) 20.
(a) Intracellular ROS intensity of Candida albicans K1 in the presence of PBS, polymer and the mixture of polymer and NAC (10 mM). n = 3 independent samples. Data are presented as mean values ± SD. (b) MIC value of polymer against Candida albicans K1 in the presence or absence of NAC.
Extended Data Fig. 5 Influence of protease, inorganic salt ions and serum on the antifungal activity of (Gly 0.8 Nap 0.2 ) 20.
(a) The MIC values for (Gly 0.8 Nap 0.2 ) 20 against Candida albicans K1 in the presence of pepsin, papain, trypsin and proteinase K at a concentration of 10 mg/mL, respectively. The changes in MIC values of (Gly 0.8 Nap 0.2 ) 20 in the presence of calcium ion (b) , magnesium ion (c) and 5% fetal bovine serum (d) , using melittin as the control.
Extended Data Fig. 6 In vivo distribution of (Gly 0.8 Nap 0.2 ) 20 .
The concentration of (Gly 0.8 Nap 0.2 ) 20 in the heart (a) , liver (b) , spleen (c) , lung (d) and kidney (e) of mice at various time points, respectively, using dye-labeled (Gly 0.8 Nap 0.2 ) 20 . n = 3 biologically independent animals. Data are presented as mean values ± SD.
Supplementary information
Supplementary information.
Materials, instrumentation and Supplementary Figs. 1–6.
Reporting Summary
Supplementary table 1 and supplementary table 2.
Antifungal activities and selectivity index of the optimal (Gly 0.8 Nap 0.2 ) 20 against drug-resistant fungi. Drug sensitivity information on the clinically isolated drug-resistant strains.
Source Data Fig. 1
GPC data for Fig. 1b and unprocessed NMR data for Fig. 1d.
Source Data Fig. 2
Fluorescence distribution curve of dye–(Gly 0.8 Nap 0.2 ) 20 and PI for Fig. 2b–d; surface zeta potential of Candida albicans K1 cells for Fig. 2e; fungal cell wall disturbance for Fig. 2f and cytoplasmic membrane depolarization of dye–(Gly 0.8 Nap 0.2 ) 20 for Fig. 2g; change in the antifungal activity of (Gly 0.8 Nap 0.2 ) 20 in the presence of phosphatidylcholine for Fig. 2h and phosphatidylethanolamine for Fig. 2i.
Source Data Fig. 3
Fluorescence intensity profiles of dye-(Gly 0.8 Nap 0.2 ) 20 and PI for Fig. 3b,c; DLS analysis for Fig. 3d; fluorescence quenching of the DNA–dye complex by the addition of (Gly 0.8 Nap 0.2 ) 20 for Fig. 3e; ultraviolet spectra for Fig. 3f and CD spectra for Fig. 3g of DNA before and after (Gly 0.8 Nap 0.2 ) 20 treatment; zeta potential of nontreated and (Gly 0.8 Nap 0.2 ) 20 -treated DNA for Fig. 3h; antifungal activity of (Gly 0.8 Nap 0.2 ) 20 for Fig. 3i.
Source Data Fig. 4
Fluorescence intensity changes of DAPI and MitoTracker Red for Fig. 4d; drug resistance test results for Fig. 4e; change of MIC values for antifungal drugs against (Gly 0.8 Nap 0.2 ) 20 -treated Candida albicans RK2911 and FLC-treated Candida albicans RK2911 for Fig. 4f; change of MIC values for (Gly 0.8 Nap 0.2 ) 20 against Candida albicans K1 in the presence of trypsin for Fig. 4g.
Source Data Fig. 5
The fungal cell burden in the infected eyeballs for Fig. 5b; clinical scores for keratitis after treatment for Fig. 5d; the fungal cell burden (log 10 (CFU g −1 )) in the infected skin for Fig. 5g.
Source Data Fig. 6
Survival rate of mouse with invasive fungal infection for Fig. 6b; fungal cell burden (log 10 (CFU g −1 )) in different organs of mice for Fig. 6c; changes in uninfected mouse body weight after (Gly 0.8 Nap 0.2 ) 20 treatment for Fig. 6e; serum biochemistry analysis on uninfected mouse for Fig. 6g.
Source Data Extended Data Fig. 2
The percentage of killed Candida albicans K1 treated with (Gly 0.8 Nap 0.2 ) 20 for Extended Data Fig. 2a.
Source Data Extended Data Fig. 3
Change in the antifungal activity of (Gly 0.8 Nap 0.2 ) 20 in the presence of exogenous fungal membrane lipid components phosphatidylglycerol (PG) for Extended Data Fig. 3a and phosphatidylinositol (PI) for Extended Data Fig. 3b.
Source Data Extended Data Fig. 4
Intracellular ROS intensity of Candida albicans K1 for Extended Data Fig. 4a. MIC value of polymer against Candida albicans K1 in the presence or absence of NAC for Extended Data Fig. 4b.
Source Data Extended Data Fig. 5
The MIC values for (Gly 0.8 Nap 0.2 ) 20 against Candida albicans K1 in the presence of pepsin, papain, trypsin and proteinase K for Extended Data Fig. 5a. The changes in MIC values of (Gly 0.8 Nap 0.2 ) 20 in the presence of calcium ion for Extended Data Fig. 5b, magnesium ion for Extended Data Fig. 5c and 5% fetal bovine serum for Extended Data Fig. 5d.
Source Data Extended Data Fig. 6
The concentration of (Gly 0.8 Nap 0.2 ) 20 in mice heart for Extended Data Fig. 6a, liver for Extended Data Fig. 6b, spleen for Extended Data Fig. 6c, lung for Extended Data Fig. 6d and kidney for Extended Data Fig. 6e.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
Reprints and permissions
About this article
Cite this article.
Zhou, M., Liu, L., Cong, Z. et al. A dual-targeting antifungal is effective against multidrug-resistant human fungal pathogens. Nat Microbiol (2024). https://doi.org/10.1038/s41564-024-01662-5
Download citation
Received : 10 October 2023
Accepted : 04 March 2024
Published : 08 April 2024
DOI : https://doi.org/10.1038/s41564-024-01662-5
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
Quick links
- Explore articles by subject
- Guide to authors
- Editorial policies
Sign up for the Nature Briefing: Translational Research newsletter — top stories in biotechnology, drug discovery and pharma.

IMAGES
VIDEO
COMMENTS
Microbiology is the study of microscopic organisms, such as bacteria, viruses, archaea, fungi and protozoa. This discipline includes fundamental research on the biochemistry, physiology, cell ...
Microbiology News. Articles and images on biochemistry research, micro-organisms, cell functions and related topics, updated daily. ... Apr. 12, 2024 — A study suggests that people with diets ...
Characterization of genes related to the efflux pump and porin in multidrug-resistant Escherichia coli strains isolated from patients with COVID-19 after secondary infection. Escherichia coli (E. coli) is a multidrug resistant opportunistic pathogen that can cause secondary bacterial infections in patients with COVID-19.This study aimed to determine the antimicrobial resistance profi...
The research was funded, in part, by the Neil and Anna Rasmussen Family Foundation. ... One catalyst for early microbiome collaboration was the Microbiology Graduate PhD Program, which recruited microbiology students to MIT and introduced them to research groups across the Institute. ... Postdoc Shaniel Bowen studies women's sexual anatomy and ...
The research should be original and include molecular aspects to generate broad interest. Papers of specialised or of preliminary and descriptive content will normally not be considered. Studies in the following sections are included: Reviews/Minireviews on all aspects ; Microbiology and Genetics ; Molecular and Cell Biology ; Metabolism and ...
The study of the human microbiome takes us back to the origins of microbiology when Antonie van Leeuwenhoek ... is the lack of diversity in research subjects, where most studies focus on western ...
Papers published in the journal cover all aspects of microbial taxonomy, phylogeny, ecology, physiology and metabolism, molecular genetics and genomics, gene regulation, viruses of prokaryotes, as well as interactions between …. View full aims & scope. Find out more about the IP. $2800. Article publishing charge.
Six Key Topics in Microbiology: 2024. in Virtual Special Issues. This collection from the FEMS journals presents the latest high-quality research in six key topic areas of microbiology that have an impact across the world. All of the FEMS journals aim to serve the microbiology community with timely and authoritative research and reviews, and by ...
Recently, the main research hotspots in microbiology include community classification and its environmental role (Bulgarelli et al., 2013; Zhang et al., ... Disease epidemiology studies examine the patterns of temporal and spatial dynamics of diseases at the population level under different environmental conditions. Research on issues such as ...
Microbiology is now fully Open Access (OA). Find out more about the transformation of the Society's founding journal and its OA future here.As the founding journal from the Microbiology Society, Microbiology brings together communities of scientists from all microbiological disciplines and from around the world. Originally Journal of General Microbiology, we have been publishing the latest ...
The current research mainly focuses on gastrointestinal diseases, while extra-intestinal diseases are also rising, such as nerve-related diseases. Although extensive correlative studies have been performed, the molecular mechanisms still need to be explored. Overall, gut microbiome research shows a multitude of challenges and great opportunities.
Journal of Medical Microbiology is the go-to interdisciplinary journal for medical, dental and veterinary microbiology, at the bench and in the clinic. It provides comprehensive coverage of medical, dental and veterinary microbiology and infectious diseases, welcoming articles ranging from laboratory research to clinical trials, including bacteriology, virology, mycology and parasitology.
The American Academy of Microbiology convened a colloquium September 5-7, 2003, in Charleston, South Carolina to discuss the central importance of microbes to life on earth, directions microbiology research will take in the 21st century, and ways to foster public literacy in this important field. Discussions centered on: The impact of microbes on the health of the planet and its inhabitants ...
microbiology, study of microorganisms, or microbes, a diverse group of generally minute simple life-forms that include bacteria, archaea, algae, fungi, protozoa, and viruses.The field is concerned with the structure, function, and classification of such organisms and with ways of both exploiting and controlling their activities.. The 17th-century discovery of living forms existing invisible to ...
Microbiology (from Ancient Greek μῑκρος (mīkros) 'small', βίος (bíos) 'life', and -λογία () 'study of') is the scientific study of microorganisms, those being of unicellular (single-celled), multicellular (consisting of complex cells), or acellular (lacking cells). Microbiology encompasses numerous sub-disciplines including virology, bacteriology, protistology, mycology ...
Microbiology Research is an international, scientific, peer-reviewed open access journal published quarterly online by MDPI ... In soils, pH stands as the main factor modulating bacterial communities' composition. However, most studies address its effects in bulk soils in natural systems, with few focusing on its effects in the rhizosphere of ...
Microorganisms matter because they affect every aspect of our lives - they are in us, on us and around us. Microbiology is the study of all living organisms that are too small to be visible with the naked eye. This includes bacteria, archaea, viruses, fungi, prions, protozoa and algae, collectively known as 'microbes'.
Microbiology is the study of microscopic organisms, such as bacteria, fungi, and protists. It also includes the study of viruses, which are not technically classified as living organisms but do contain genetic material. Microbiology research encompasses all aspects of these microorganisms such as their behavior, evolution, ecology, biochemistry ...
The aims of this study were to determine which factors are considered by microbiology and immunology educators in the United States before they incorporate a new and emerging topic into their curriculum, define the microbiology and immunology topics considered new and emerging at a single snapshot in time (2022), and explore best practices of ...
Advances in microbiology are largely driven by improvements in technology. The awe-inspiring size of the experiments performed today — such as studies of entire microbial communities — is the ...
Welcome to the wonderful world of microbiology! Yay! So. What is microbiology? If we break the word down it translates to "the study of small life," where the small life refers to microorganisms or microbes. But who are the microbes? And how small are they? Generally microbes can be divided in to two categories: the cellular microbes (or organisms) and the acellular microbes (or agents ...
This study shows that dual-targeting antifungal compounds may be effective in combating drug-resistant fungal pathogens and mitigating fungal resistance. You have full access to this article via ...